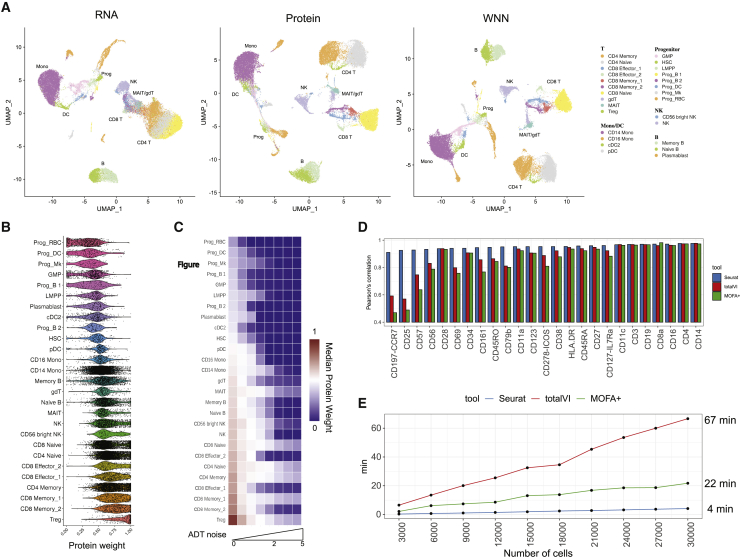
Figure 2.
Benchmarking and robustness analysis for WNN integration
(A) Analysis of a CITE-seq dataset of human bone marrow mononuclear cells and 25 surface proteins. UMAP visualizations are computed using RNA, protein, or WNN analysis. Cell annotations are derived from WNN analysis and reveal heterogeneity within T cells and progenitors that cannot be discovered by either modality independently. Granular annotations, which more clearly indicate subpar performance when analyzing only one modality, are shown in Figure S2.
(B) Single-cell protein modality weights. Progenitor populations all receive low protein weights, whereas T cell populations receive high protein modality weights, consistent with the composition of the antibody panel that was tailored for differentiated cell types.
(C) To test the robustness of WNN, we added increasing amounts of Gaussian noise to the protein data. Protein weights decrease to 0 in all cell types as noise levels increase.
(D and E) Benchmarking WNN against totalVI and MOFA+. (D) The integrated latent space defined by WNN most accurately reconstructs expression levels for 25 proteins. (E) WNN analysis exhibits improved runtimes compared to competing methods. Additional benchmarking analyses in Figure S2.
See also Figure S3.