Figure 4.
Multimodal biomarkers of immune cell states
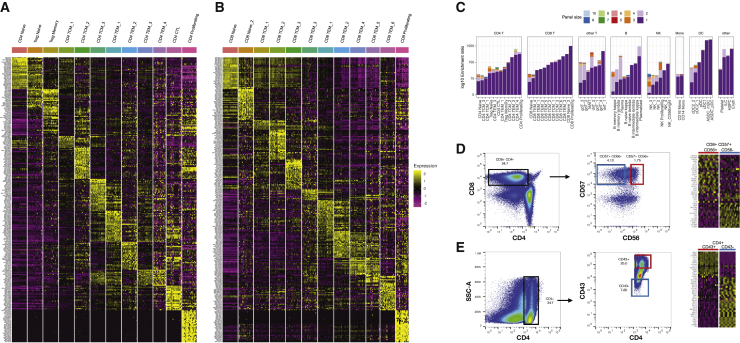
(A) Heatmap of CD4+ T cell states. Markers include the best RNA and protein features identified by differential expression (DE). Heatmap displays pseudobulk averages where cells are grouped by cell type, donor, and vaccination time point and demonstrates that markers do not vary across different PBMC samples.
(B) Same as in (A) but for CD8+ T cell states. Additional heatmaps are shown in Figure S4.
(C) For each of our 57 clusters, we calculated the optimal surface marker enrichment panels based on our CITE-seq data. Bar plots show the ability of the panels to enrich for each cell type in silico. The composition of each panel is shown in Table S2.
(D) Validation of predicted marker panels for the CD8_TEM_5 cluster. We sorted cells based on the marker panels identified in (C), and performed bulk RNA-seq. Each column represents a replicate bulk RNA-seq profile. Heatmap is ordered by genes expected to be DE based on our CITE-seq dataset and are validated by bulk RNA-seq.
(E) Same as in (D) but for CD4 CTL cells.