Abstract
Microbial natural products (NPs) are of paramount importance in human medicine, animal health and plant crop protection. Large-scale microbial genome and metagenomic mining has revealed tremendous biosynthetic potential to produce new NPs. However a majority of NP biosynthetic gene clusters (BGCs) are functionally inaccessible under standard laboratory conditions. BGC refactoring and heterologous expression provide a promising synthetic biology approach to NP discovery, yield optimization and combinatorial biosynthesis studies. In this review, we summarize the recent advances pertaining to the heterologous production of bacterial and fungal NPs, with an emphasis on next-generation transcriptional regulatory modules, novel BGC refactoring techniques and optimized heterologous hosts.
Introduction
Microbial natural products (NPs) and their synthetic derivatives play a significant role in drug discovery and development as a result of their rich chemical diversity and propensity to exhibit bioactivity [1,2]. Historically, bacterial or fungal NPs as well as their derivatives have been widely used in human health and agriculture [1]. The NP discovery field enjoyed a ‘golden era’ ushered in by the Waksman platform based on fermentation-coupled bioactivity screening [3]. However, known compounds are now increasingly re-discovered when using this traditional screening platform. With the advances in genome sequencing methods, bioinformatics analysis algorithms and synthetic biology tools, we are currently witnessing a renaissance of NP discovery in the post-genomic era [4–7]. The development of high-throughput and low-cost sequencing technology has accelerated the exploration of bacterial and fungal genomes, thus resulting in an exponential growth of microbial genomic sequencing data [8]. Advanced computational tools, such as antiSMASH [9] and PRISM [10], have been developed for large-scale prediction of NP biosynthetic gene clusters (BGCs) and their encoded chemical structures [11]. Repositories for BGCs with characterized metabolites, such as MIBiG 2.0 (Minimum Information about a Biosynthetic Gene cluster) and IMG-ABC v.5.0 (Integrated Microbial Genomes Atlas of Biosynthetic gene Clusters), have been established to further strengthen in silico analysis of novel BGCs [12,13]. Finally, innovative synthetic biology and metabolic engineering tools have greatly accelerated BGC cloning and refactoring as well as host genetic engineering [14–16].
Many microorganisms, especially actinomycetes, cyanobacteria and myxobacteria, contain an order of magnitude more BGCs than are expressed in the laboratory [2,17]. BGC refactoring by replacing natural promoters with constitutive or readily inducible promoters and heterologous expression in a panel of optimized heterologous expression strains has the potential to provide access to the chemical diversity encoded by silent BGCs for a number of reasons [18–20]. Versatile surrogate heterologous hosts and general synthetic biology toolboxes are broadly applicable to genetic manipulation of cloned BGCs from diverse sources, such as the human microbiome [21] and marine ecosystems [22]. New genetic tools are not required to interrogate native BGCs from each new species or genus. Furthermore, heterologous expression enables culture-independent characterization of bioactive small molecules from obligate symbionts and environmental DNA (e.g., soil metagenomes) [22–24].
This review aims to provide insight into general BGC refactoring strategies for heterologous production of bioactive NPs for next-generation drug discovery. New designs for orthogonal transcriptional regulatory modules are discussed, which provide a wide variety of species-selective toolkits for high-efficiency BGC promoter engineering. We further describe a panel of widely used BGC refactoring approaches for successful heterologous reconstitution of various NPs. Lastly, we highlight recent studies on genetic engineering strategies to generate powerful heterologous hosts for use in the isolation of new compounds as well as the overproduction of important pharmaceutically active NPs.
Next-generation transcriptional regulatory modules for gene expression control
For efficient BGC refactoring, a panel of orthogonal transcriptional regulatory elements including promoters, ribosomal binding sites (RBSs), terminators and protein degradation tags, is indispensable [25–28]. The synthetic biology toolboxes are regulatable and enable coordinated expression of refactored BGCs [27–29]. Considering that promoters are responsible for the first stage of gene expression, they are critical for activation and functional optimization of BGCs in heterologous hosts. Through systematic exchanges between native transcriptionally silent promoters and constitutive promoters, promoter engineering provides a universal strategy to disrupt native transcriptional regulation networks and subsequently activate silent BGCs. In the last two decades, a panel of native and synthetic promoter libraries has been constructed for efficient transcriptional control of NP biosynthetic pathways [26,27]. However, these well-characterized regulatory modules possess some limitations to be overcome, including low sequence divergence and narrow applications in only certain microorganisms. Here, we discuss a panel of new design concepts to generate next-generation regulatory elements for gene expression control across different genetic backgrounds and various growth conditions.
Orthogonal regulatory elements by randomizing sequences in both promoter and RBS regions
Compared to native promoter libraries, synthetic promoter libraries can efficiently avoid host perturbation due to the removal of unessential DNA regions. However, almost all synthetic bacterial promoter libraries have been generated by randomization of the spacer between −10 and −35 regions, which decreases the degree of sequence divergence and possibly leads to homologous recombination of promoters in refactored BGCs [26]. Recently, Ji et al. presented a new design concept of synthetic promoter libraries in a model actinomycete, Streptomyces albus J1074 (Figure 1a) [30]. Based on a series of characterized constitutive promoters, the regulatory sequences including both promoter and RBS regions were completely randomized by only partially fixing −10/−35 regions and the Shine-Dalgarno (SD) sequence in the RBS region. Using a nonribosomal peptide synthetase (NRPS) that produces the blue pigment indigoidine as a reporter, a large pool of regulatory sequences with strong, medium or weak transcriptional activities was constructed by monitoring indigoidine production. Theoretically, these regulatory elements should be highly orthogonal, which can significantly facilitate multiplex promoter engineering of multiple operon-containing BGCs in actinomycetes. To demonstrate the utility of these synthetic regulatory cassettes in engineering BGCs, the actinorhodin (ACT) BGC from Streptomyces coelicolor was refactored by replacing the seven native promoters with the four strong regulatory cassettes. While the native ACT BGC was silent in minimal media, the engineered ACT BGC was successfully heterologously expressed in S. albus J1074 [30]. Furthermore, this completely randomized design mode of 5’ regulatory sequences could be applicable to the construction of synthetic promoter libraries in other model heterologous expression strains, such as Myxococcus xanthus DK1622 and Burkholderia sp. DSM7029 [31,32].
Figure 1. Next-generation transcriptional regulatory modules for gene expression control.
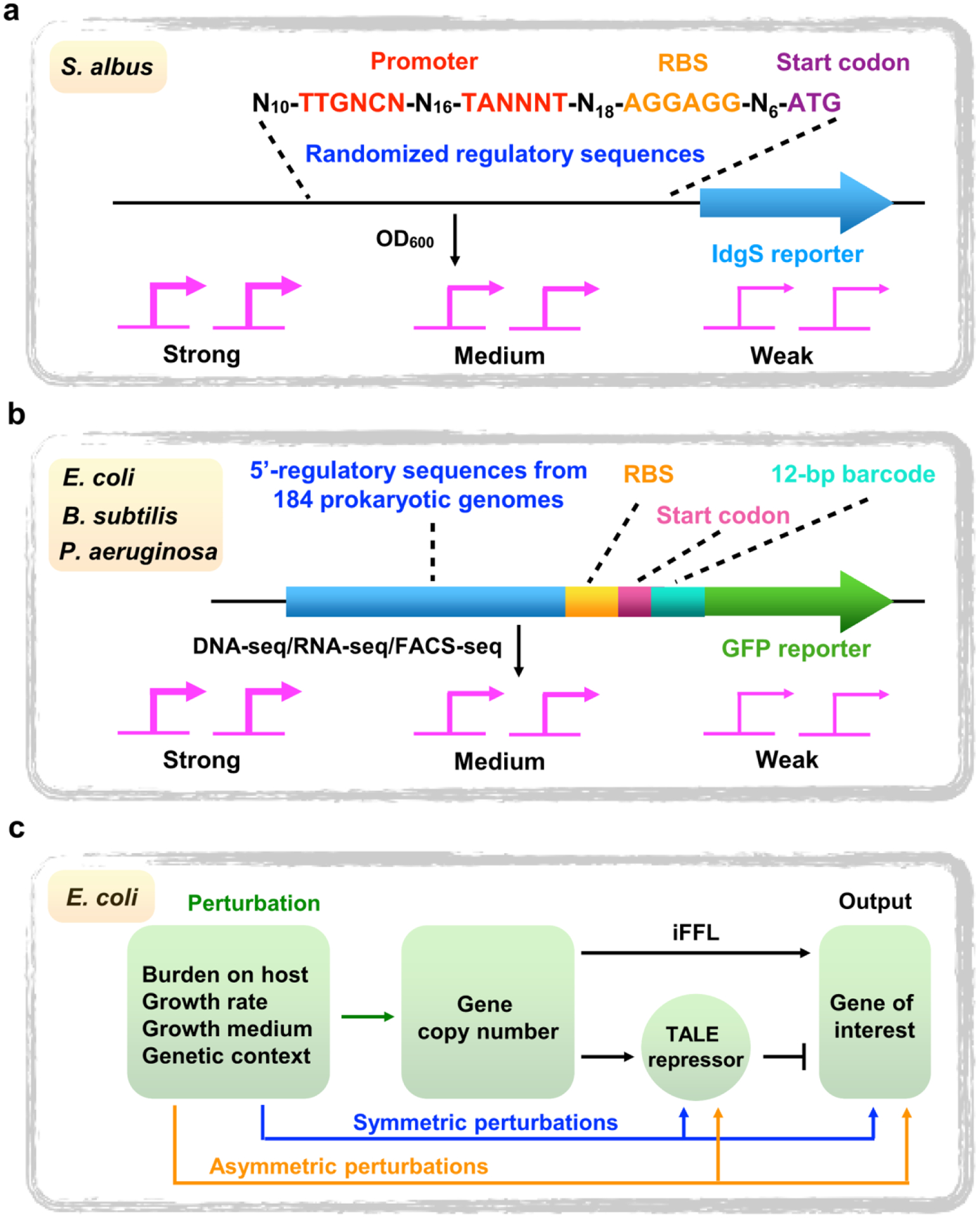
a) Completely randomized regulatory sequences in both promoter and RBS regions in S. albus. IdgS is a nonribosomal peptide synthetase (NRPS), which produces the blue pigment indigoidine; b) Metagenomic mining of natural 5’ regulatory elements from diverse bacteria; c) iFFL-stabilized promoters in E. coli. iFFL, incoherent FeedForward Loop. TALE, Transcription-Activator Like Effector.
Metagenomic mining of natural 5’ regulatory elements with universal host ranges
A large proportion of microbial bioactive NPs have arisen from examining the same limited number of bacterial taxa (e.g., actinomycetes, bacilli, cyanobacteria, myxobacteria and filamentous fungi). In recent years this has led to the frequent rediscovery of known NPs [1,2]. Therefore, focusing on untapped producers is an obvious advantage in finding new classes of antibiotics and other pharmaceutical compounds [4]. In recent years, new sources of NPs have been explored, including previously understudied soil bacteria as well as human and nematode microbiomes [4,33]. In this regard, it is critical to establish a panel of promoter libraries with universal host ranges for BGC refactoring in underexplored bacterial taxa. In 2018, Johns et al. mined 184 microbial genomes to expand the phylogenetic breadth of promoters, thus generating a diverse library of natural 5’ regulatory sequences from Actinobacteria, Archaea, Bacterioidetes, Cyanobacteria, Fimicutes, Proteobacteria and Spirochetes (Figure 1b) [34]. By assembling the putative 5’ regulatory regions into species-specific vectors, both transcriptional and translational levels of large-scale regulatory elements were systematically quantified using GFP as a reporter across different bacterial species and growth conditions. A common subset of regulatory elements with varying sequence composition and orthogonal host ranges, was identified [34]. This data set expands the repertoire of natural promoters, which represents a rich resource for tuning gene expression across a wide range of bacteria. Of note, these natural regulatory sequences were tested only in Escherichia coli, Bacillus subtilis and Pseudomonas aeruginosa; confirmation of their applicability to refactor BGCs in common NP-producing bacteria (e.g., streptomycetes and myxobacteria) is still required.
Engineered promoters with constant gene expression at any copy number
Gene expression levels are often affected by a variety of conditions, including growth conditions, growth phases and metabolic burden. Refactoring BGCs-of-interest using stabilized promoters is an obvious advantage when activating silent BGCs in diverse heterologous hosts. Using transcription-activator like effectors (TALEs)-based incoherent feedforward loop (iFFL), Segall-Shapiro et al. developed a series of engineered promoters with constant expression levels at any copy numbers in E. coli (Figure 1c) [35]. The robustness of these promoters was demonstrated by near-identical expression strengths in different plasmid backbones or genome locations. For example, the titers of deoxychromoviridans are nearly identical when transferring its BGC from a high-copy plasmid to the host genome. Interestingly, iFFL-stabilized promoters enable the design of metabolic pathways that are resistant to changes in genome mutations, growth conditions or other stressors [35].
Novel BGC refactoring strategies for activation of silent BGCs or yield optimization
For heterologous reconstitution of NP biosynthetic pathways, BGC cloning is the first challenging step due to large cluster size and repetitive regions. A wide variety of BGC cloning methods have been developed over the years, including cosmid/fosmid/BAC/FAC library-based methods, direct cloning methods and bottom-up assembly methods. Interested readers are referred to the recently published reviews [18,19,36]. Of note, it is estimated that about 90% of native BGCs are not expressed or are partially transcriptional under standard laboratory conditions [17]. Here, we therefore focus on new BGC refactoring strategies for activation of silent BGCs or pathway optimization (Figure 2).
Figure 2. Refactoring biosynthetic gene clusters for novel NP discovery or yield optimization of clinically relevant drugs.
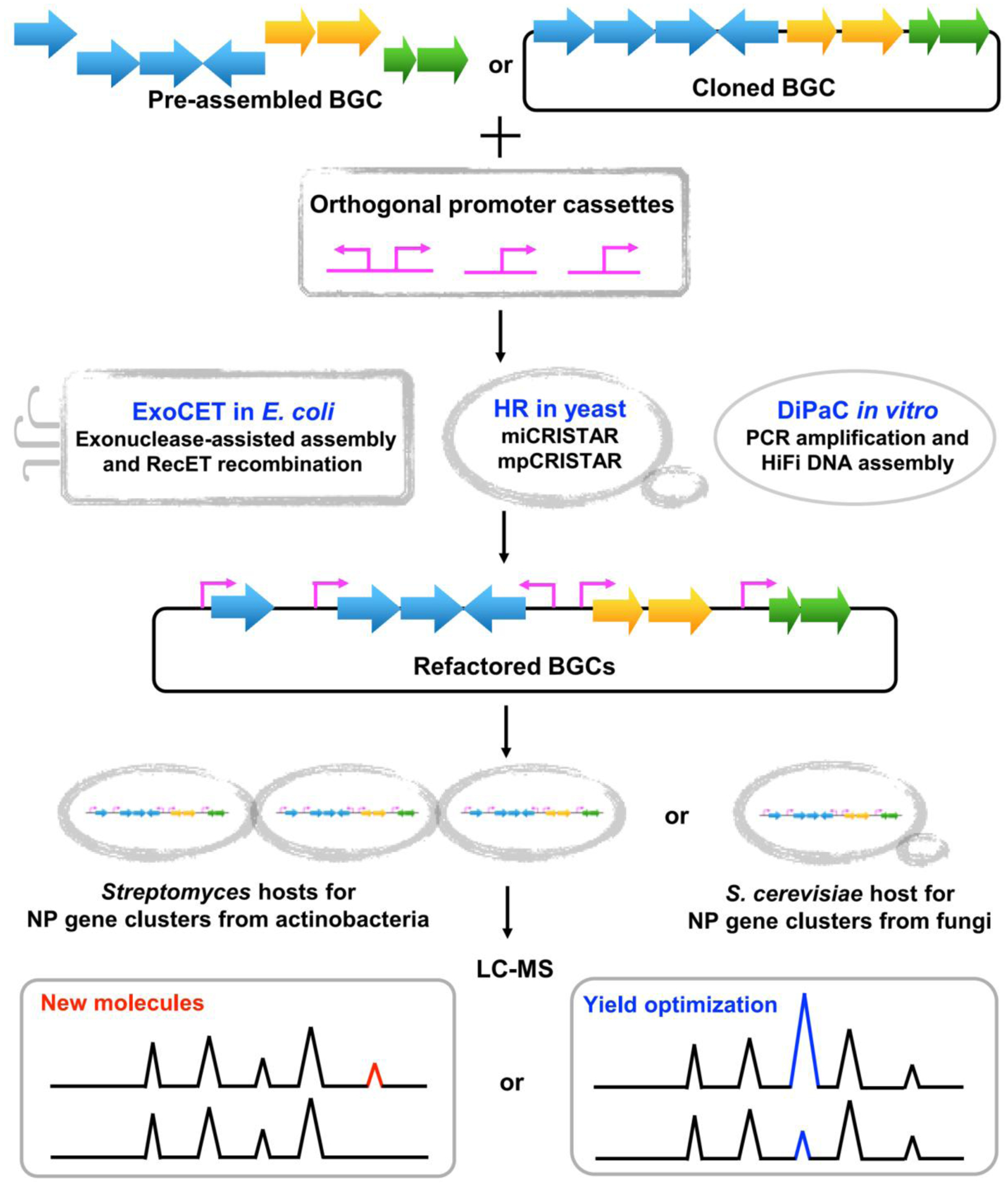
BGC, biosynthetic gene cluster; HR, homologous recombination; NP, natural product; LC-MS, liquid chromatography-mass spectrometry.
Generally, BGC refactoring involves genetic manipulation of entire cloned BGCs and simultaneous modification when assembling target BGCs (Figure 2). Based on powerful yeast homologous recombination (YHR), a series of in vivo BGC editing methods for multiplexed promoter engineering have been developed with simultaneous replacement of up to eight promoters with high efficiency, including mCRISTAR (multiplexed CRISPR-based Transformation-Associated Recombination (TAR)), miCRISTAR (multiplexed in vitro CRISPR-based TAR) and mpCRISTAR (multiple plasmid-based CRISPR-based TAR) [20,37–39]. For instance, miCRISTAR-mediated fast activation of a silent BGC led to the discovery of two antitumor sesterterpenes atolypene A and B [38]. Recently, Harvey et al. achieved assembly of up to 14 unique DNA fragments with high efficiency using YHR. Their yeast heterologous expression platform (HEx) has been successfully used to reconstruct 41 fungal BGCs from diverse fungal species, 22 of which produced detectable compounds [27]. In another study, YHR-mediated reconstitution of a riboswitch-controlled pathway achieved a 120-fold increase in bottromycin production [40]. In contrast to YHR-based strategies, a series of in vitro BGC assembly methods have been established to reconstruct target BGCs for diverse applications. For example, an innovative DNA assembly method, ExoCET (Exonuclease Combined with RecET recombination), was developed for refactoring large, multi-operon BGCs in E. coli [41,42]. The 79-kb spinosad BGC with seven artificial operons under the control of strong constitutive promoters achieved 328-fold enhanced spinosad production compared to the native BGC [40]. Finally, using long amplification PCR and HiFi DNA in vitro assembly, an emerging synthetic biology approach, DiPaC (Direct Pathway Cloning), has been established for fast BGC cloning and refactoring [43,44]. Removing predicted transcriptional terminators when assembling the cyanobacterial hapalosin BGC (23 kb) led to successful expression of the hapalosin BGC in E. coli [44]. These newly developed DNA refactoring methods provide a time- and cost-efficient platform to discover bioactive NPs at an unprecedented scale from either metagenomes or cultured microorganisms. Particularly, when multiple promoters are required to refactor BGCs that have been cloned into a vector, a YHR-based strategy is often preferred due to its efficiency and accuracy compared to other BGC refactoring strategies [20, 38].
Genetic engineering strategies for the construction of powerful heterologous hosts
A good heterologous host must grow quickly, be genetically tractable and be able to supply all precursors and co-factors for biosynthesis of exogenous NPs [18]. In the last two decades, a series of excellent heterologous hosts, including Gram-positive bacteria, Gram-negative bacteria and fungi, were developed for the discovery of novel NPs from diverse sources [18,45,46]. Of note, screening phylogenetically diverse strains may significantly enhance the success rate of BGC heterologous expression studies [45,47]. For instance, our group examined 38 diverse Streptomyces hosts for heterologous expression of 97 environmental DNA (eDNA) cosmid clones containing minimal type II polyketide synthases (PKS), and successfully identified a new tricyclic polyene metatricycloene in Streptomyces albus, which exhibited the best innate ability for BGC heterologous expression [47]. In another study, Wang et al. developed chassis-independent recombinase-assisted genome engineering (CRAGE) for high-efficient integration of complex BGC constructs in 25 diverse gamma-Proteobacteria species, thus activating six BGCs from Photorhabdus luminescens and generating 22 diverse compounds [48]. Interested readers are referred to the recently published reviews [18,45,46,49]. Here, we will focus on general genetic strategies for the optimization of heterologous expression strains, particularly Streptomyces spp. and S. cerevisiae, which can be extended to engineer other bacterial or fungal species.
Diverse Streptomyces spp. are widely used for heterologous expression of BGCs from actinobacteria or other bacteria with high-GC content genomes [46,50]. These surrogate hosts have been combinatorially optimized by a series of universal genetic strategies for enhanced production titers of specific NPs (Figure 3a). Deletion of non-essential BGCs can increase the supply of primary metabolic precursors and simultaneously simplify the host’s secondary metabolome for downstream analysis [51,52]. Multi-copy amplification of target BGCs can be achieved by introducing multiple site-specific integration sites (i.e. phiC31 attB sites) into the host genome in advance, thus resulting in increased productivity of a target compound [52–54]. Empirically identified mutations with positive effects on NP biosynthesis in rpoB and rpsL (encoding the β-subunits of RNA polymerase and ribosomal protein S12, respectively) are often introduced [55]. Reprogramming host regulatory networks by overexpressing positive global regulators and/or deleting negative global regulatory genes has also been leveraged [56]. Finally, introduction of efflux pumps, such as LmrA and MdfA, can reduce end-product toxicity and increase extraction yields of target NPs [56].
Figure 3. Genetic engineering strategies for optimized microbial strain construction to facilitate BGC heterologous expression.
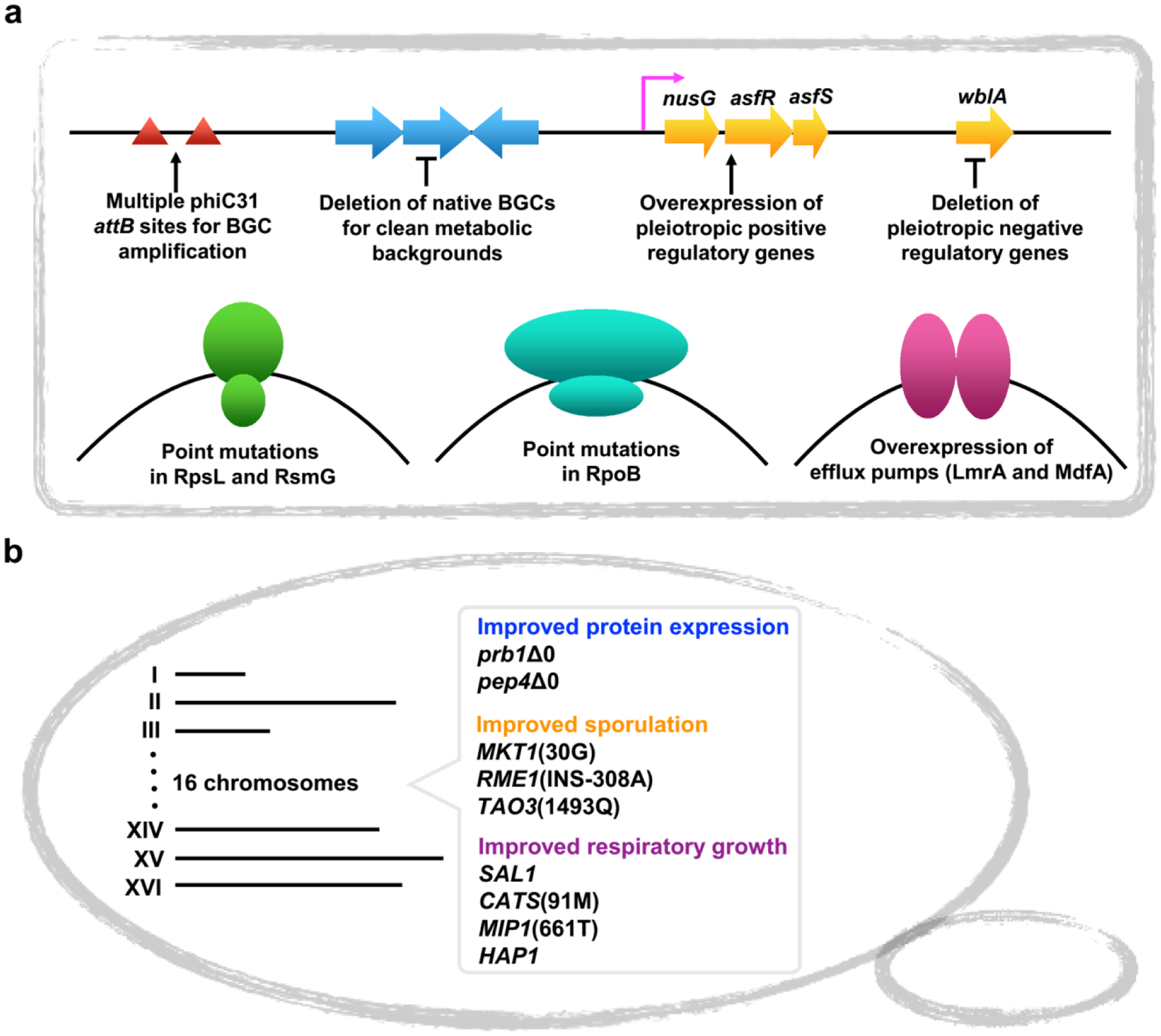
a) General Streptomyces host modification strategies; b) S. cerevisiae host modifications with improved growth and expression phenotypes.
Filamentous fungi are regarded as rich sources for discovery of bioactive NPs [6]. S. cerevisiae exhibits many advantages for heterologous expression of BGCs from filamentous fungi due to its well-studied metabolism and extensive genetic toolbox [18,49]. Recently, a heterologous expression platform was developed for rapid and scalable expression of completely refactored fungal BGCs in S. cerevisiae [27]. The defects of well-characterized S. cerevisiae S288c-derived yeast strains were systematically optimized (Figure 3b). Mitochondrial stability and sporulation ability are first increased based on a series of specific genetic modifications. By deleting the vacuolar protease encoding genes, heterologous protein production is improved. Finally, a series of genes for essential posttranslational modification enzymes are integrated. Compared to common laboratory strains, the newly engineered S. cerevisiae strain with improved growth and expression phenotypes provides an improved heterologous expression strain for fungal NP discovery [27].
Concluding remarks and future perspectives
Microbial NPs play an instrumental role in the discovery of antibacterials, anticancer agents and agrochemicals [4,57]. With the continuing development of genomics and synthetic biotechnology, activation of silent BGCs in heterologous expression strains has been used extensively as a promising avenue to access bioactive NPs [18,19]. Expanding the spectrum of heterologous hosts and the host range of promoter elements will likely enable the more facile discovery of novel BGCs from underexplored species and metagenomic sequence datasets. The enabling technologies discussed in this review and future developments are very likely to facilitate the discovery of novel chemical scaffolds and perhaps eventually help to expand our repertoire of therapeutic agents.
Acknowledgement
This work was supported by a grant from NIH (5R35GM122559).
Footnotes
Conflict of interest statement
The authors declare no competing financial interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Newman DJ, Cragg GM: Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 2016, 79:629–661. [DOI] [PubMed] [Google Scholar]
- 2.Katz L, Baltz RH: Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol 2016, 43:155–176. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K: Antibiotics: Recover the lost art of drug discovery. Nature 2012, 485:439–440. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K: The science of antibiotic discovery. Cell 2020, 181:29–45. [DOI] [PubMed] [Google Scholar]; •• The author summarizes the history and challenges of natural product discovery, and discusses an emerging new platform for antibiotic discovery from untapped producers.
- 5.Kalkreuter E, Pan GH, Cepeda AJ, Shen B: Targeting bacterial genomes for natural product discovery. Trends Pharmacol Sci 2020, 41:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller NP: Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol 2019, 17:167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen B: A new golden age of natural products drug discovery. Cell 2015, 163:1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Katta HY, Mojica A, Chen IMA, Kyrpides NC, Reddy TBK: Genomes OnLine Database (GOLD) v.7: updates and new features. Nucleic Acids Res 2019, 47:D649–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T: antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 2019, 47:W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skinnider MA, Merwin NJ, Johnston CW, Magarvey NA: PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res 2017, 45:W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziemert N, Alanjary M, Weber T: The evolution of genome mining in microbes - a review. Nat Prod Rep 2016, 33:988–1005. [DOI] [PubMed] [Google Scholar]
- 12.Kautsar SA, Blin K, Shaw S, Navarro-Munoz JC, Terlouw BR, van der Hooft JJJ, van Santen JA, Tracanna V, Suarez Duran HG, Pascal Andreu V, et al. : MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res 2020, 48:D454–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palaniappan K, Chen IA, Chu K, Ratner A, Seshadri R, Kyrpides NC, Ivanova NN, Mouncey NJ: IMG-ABC v.5.0: an update to the IMG/Atlas of biosynthetic gene clusters knowledgebase. Nucleic Acids Res 2020, 48:D422–D430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbasi MN, Fu J, Bian X, Wang H, Zhang Y, Li A: Recombineering for genetic engineering of natural product biosynthetic pathways. Trends Biotechnol 2020. 10.1016/j.tibtech.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Skellam E: Strategies for engineering natural product biosynthesis in fungi. Trends Biotechnol 2019, 37:416–427. [DOI] [PubMed] [Google Scholar]
- 16.Palazzotto E, Tong Y, Lee SY, Weber T: Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol Adv 2019, 37:107366. [DOI] [PubMed] [Google Scholar]
- 17.Rutledge PJ, Challis GL: Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 2015, 13:509–523. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JJ, Tang XY, Moore BS: Genetic platforms for heterologous expression of microbial natural products. Nat Prod Rep 2019, 36:1313–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors highlight a panel of genetic platforms to identify, interrogate and engineer BGCs, including BGC cloning and refactoring techniques as well as diverse heterologous hosts.
- 19.Huo LJ, Hug JJ, Fu CZ, Bian XY, Zhang YM, Muller R: Heterologous expression of bacterial natural product biosynthetic pathways. Nat Prod Rep 2019, 36:1412–1436. [DOI] [PubMed] [Google Scholar]; • The authors summarize heterologous production of bacterial natural products with an emphasis on novel chemistry since 2013.
- 20.Montiel D, Kang HS, Chang FY, Charlop-Powers Z, Brady SF: Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc Natl Acad Sci U S A 2015, 112:8953–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LL, Ravichandran V, Yin YL, Yin J, Zhang YM: Natural products from mammalian gut microbiota. Trends Biotechnol 2019, 37:492–504. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal V, Blanton JM, Podell S, Taton A, Schorn MA, Busch J, Lin Z, Schmidt EW, Jensen PR, Paul VJ, et al. : Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat Chem Biol 2017, 13:537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlop-Powers Z, Milshteyn A, Brady SF: Metagenomic small molecule discovery methods. Curr Opin Microbiol 2014, 19:70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hover BM, Kim SH, Katz M, Charlop-Powers Z, Owen JG, Ternei MA, Maniko J, Estrela AB, Molina H, Park S, et al. : Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat Microbiol 2018, 3:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horball L, Siegl T, Luzhetskyy A: A set of synthetic versatile genetic control elements for the efficient expression of genes in actinobacteria. Sci Rep 2018, 8:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myronovskyi M, Luzhetskyy A: Native and engineered promoters in natural product discovery. Nat Prod Rep 2016, 33:1006–1019. [DOI] [PubMed] [Google Scholar]
- 27.Harvey CJB, Tang MC, Schlecht U, Horecka J, Fischer CR, Lin HC, Li J, Naughton B, Cherry J, Miranda M, et al. : HEx: A heterologous expression platform for the discovery of fungal natural products. Sci Adv 2018, 4:eaar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A high-throughput heterologous expression platform to unlock fungal natural product biosynthetic potentials at an unprecedented scale, including universal genetic parts, improved yeast host and powerful DNA assembly strategy.
- 28.Bai C, Zhang Y, Zhao X, Hu Y, Xiang S, Miao J, Lou C, Zhang L: Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc Natl Acad Sci U S A 2015, 112:12181–12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horbal L, Marques F, Nadmid S, Mendes MV, Luzhetskyy A: Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab Eng 2018, 49:299–315. [DOI] [PubMed] [Google Scholar]
- 30.Ji CH, Kim JP, Kang HS: Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth Biol 2018, 7:1946–1955. [DOI] [PubMed] [Google Scholar]; • A completely randomized design concept for the construction of synthetic promoter libraries in bacteria.
- 31.Sucipto H, Pogorevc D, Luxenburger E, Wenzel SC, Muller R: Heterologous production of myxobacterial alpha-pyrone antibiotics in Myxococcus xanthus. Metab Eng 2017, 44:160–170. [DOI] [PubMed] [Google Scholar]
- 32.Bian XY, Tang B, Yu YC, Tu Q, Gross F, Wang HL, Li AY, Shen YM, Li YZ, Stewart AF, et al. : Heterologous production and yield improvement of epothilones in Burkholderiales strain DSM 7029. ACS Chem Biol 2017, 12:1805–1812. [DOI] [PubMed] [Google Scholar]
- 33.Niu GQ, Li WL: Next-generation drug discovery to combat antimicrobial resistance. Trends Biochem Sci 2019, 44:961–972. [DOI] [PubMed] [Google Scholar]
- 34.Johns NI, Gomes ALC, Yim SS, Yang A, Blazejewski T, Smillie CS, Smith MB, Alm EJ, Kosuri S, Wang HH: Metagenomic mining of regulatory elements enables programmable species-selective gene expression. Nat Methods 2018, 15:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Metagenomic mining of a series of orthogonal transcriptional regulatory elements with universal and orthogonal host ranges from diverse bacterial species.
- 35.Segall-Shapiro TH, Sontag ED, Voigt CA: Engineered promoters enable constant gene expression at any copy number in bacteria. Nat Biotechnol 2018, 36:352–358. [DOI] [PubMed] [Google Scholar]
- 36.Xu M, Wright GD: Heterologous expression-facilitated natural products’ discovery in actinomycetes. J Ind Microbiol Biotechnol 2019, 46:415–431. [DOI] [PubMed] [Google Scholar]
- 37.Kim H, Ji CH, Je HW, Kim JP, Kang HS: mpCRISTAR: Multiple plasmid approach for CRISPR/Cas9 and TAR-mediated multiplexed refactoring of natural product biosynthetic gene clusters. ACS Synth Biol 2020, 9:175–180. [DOI] [PubMed] [Google Scholar]
- 38.Kim SH, Lu W, Ahmadi MK, Montiel D, Ternei MA, Brady SF: Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach. ACS Synth Biol 2019, 8:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A high-efficient BGC editing method combining in vitro CRISPR-Cas9 system and yeast homologous recombination.
- 39.Kang HS, Charlop-Powers Z, Brady SF: Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth Biol 2016, 5:1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eyles TH, Vior NM, Truman AW: Rapid and robust yeast-mediated pathway refactoring generates multiple new bottromycin-related metabolites. ACS Synth Biol 2018, 7:1211–1218. [DOI] [PubMed] [Google Scholar]
- 41.Song CY, Luan J, Cui QW, Duan QY, Li Z, Gao YS, Li RJ, Li AY, Shen YM, Li YZ, et al. : Enhanced heterologous spinosad production from a 79-kb synthetic multioperon assembly. ACS Synth Biol 2019, 8:137–147. [DOI] [PubMed] [Google Scholar]
- 42.Wang HL, Li Z, Jia RN, Yin J, Li AY, Xia LQ, Yin YL, Muller R, Fu J, Stewart AF, et al. : ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res 2018, 46:2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greunke C, Duell ER, D’Agostino PM, Glockle A, Lamm K, Gulder TAM: Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential. Metab Eng 2018, 47:334–345. [DOI] [PubMed] [Google Scholar]; • A high-efficient BGC cloning and refactoring method combining long-amplicon PCR and HiFi DNA assembly.
- 44.D’Agostino PM, Gulder TAM: Direct pathway cloning combined with sequence- and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression. ACS Synth Biol 2018, 7:1702–1708. [DOI] [PubMed] [Google Scholar]
- 45.Ke J, Yoshikuni Y: Multi-chassis engineering for heterologous production of microbial natural products. Curr Opin Biotechnol 2020, 62:88–97. [DOI] [PubMed] [Google Scholar]
- 46.Myronovskyi M, Luzhetskyy A: Heterologous production of small molecules in the optimized Streptomyces hosts. Nat Prod Rep 2019, 36:1281–1294. [DOI] [PubMed] [Google Scholar]; • The authors highlight a panel of universal Streptomyces hosts for heterologous expression of natural products from actinobacteria and other bacteria with high-GC content genomes.
- 47.Iqbal HA, Low-Beinart L, Obiajulu JU, Brady SF: Natural product discovery through improved functional metagenomics in Streptomyces. J Am Chem Soc 2016, 138:9341–9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang GY, Zhao ZY, Ke J, Engel Y, Shi YM, Robinson D, Bingol K, Zhang ZY, Bowen B, Louie K, et al. : CRAGE enables rapid activation of biosynthetic gene clusters in undomesticated bacteria. Nat Microbiol 2019, 4:2498–2510. [DOI] [PubMed] [Google Scholar]
- 49.Bond C, Tang Y, Li L: Saccharomyces cerevisiae as a tool for mining, studying and engineering fungal polyketide synthases. Fungal Genet Biol 2016, 89:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu R, Deng ZX, Liu TG: Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab Eng 2018, 50:74–84. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed Y, Rebets Y, Estevez MR, Zapp J, Myronovskyi M, Luzhetskyy A: Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb Cell Fact 2020, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myronovskyi M, Rosenkranzer B, Nadmid S, Pujic P, Normand P, Luzhetskyy A: Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab Eng 2018, 49:316–324. [DOI] [PubMed] [Google Scholar]; •• A representative study for developing powerful Streptomyces chassis strains by systematically deleting non-essential native BGCs and introducing multiple site-specific integration sites.
- 53.Li L, Zheng GS, Chen J, Ge M, Jiang WH, Lu YH: Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab Eng 2017, 40:80–92. [DOI] [PubMed] [Google Scholar]
- 54.Pei ZF, Yang MJ, Li L, Jian XH, Yin Y, Li D, Pan HX, Lu Y, Jiang W, Tang GL: Directed production of aurantizolicin and new members based on a YM-216391 biosynthetic system. Org Biomol Chem 2018, 16:9373–9376. [DOI] [PubMed] [Google Scholar]
- 55.Gomez-Escribano JP, Bibb MJ: Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 2011, 4:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng Q, Gao G, Lu J, Long Q, Chen X, Zhang F, Xu M, Liu K, Wang Y, Deng Z, et al. : Engineered Streptomyces lividans strains for optimal identification and expression of cryptic biosynthetic gene clusters. Front Microbiol 2018, 9:3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hug JJ, Krug D, Muller R: Bacteria as genetically programmable producers of bioactive natural products. Nat Rev Chem 2020. 10.1038/s41570-020-0176-1. [DOI] [PubMed] [Google Scholar]


