Fig. 1.
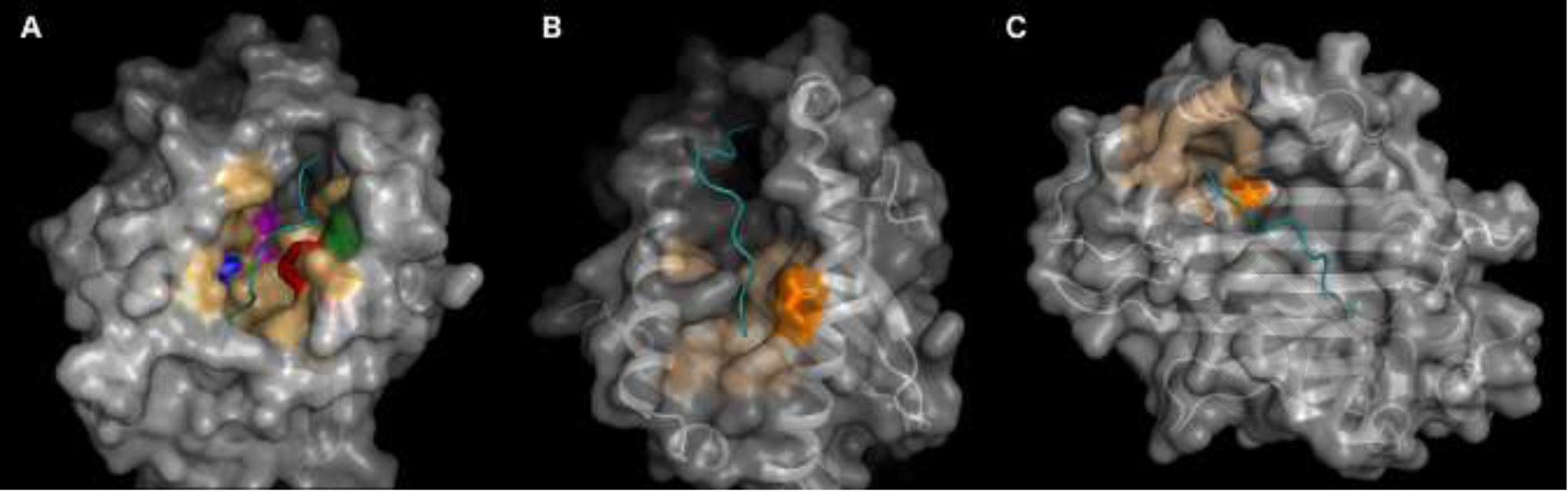
Locations of the significant residues. The 3-dimensional structure of each HLA Class I molecule is shown, highlighting the significant residues and pockets in which they participate. In all images the HLA protein is shown with a carbon backbone overlayed with the surface. The peptide presented by the HLA molecule is shown in cyan, and the non-significant HLA pocket residues are shown in light orange. The significant residue(s) for each HLA molecule is shown in a bold color and with the side chains as sticks. Beta-2 microglobulin is present as part of each crystal structure and has been hidden. A) HLA-A, based on an HLA-A*01:01 crystal structure (Protein Data Bank (PDB): 6AT9), has been mutated to show a Valine residue at position 152. Each significant residue is shown separately: A9F is blue, A97I is magenta, A152V is green and A156R is red. B) HLA-B, based on a HLA-B*27:05 crystal structure (PDB ID: 2BSS) shows B163E as orange. C) HLA-C, based on a HLAC*06:02 crystal structure (PDB ID: 5 W67), shows C116S also as orange. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
