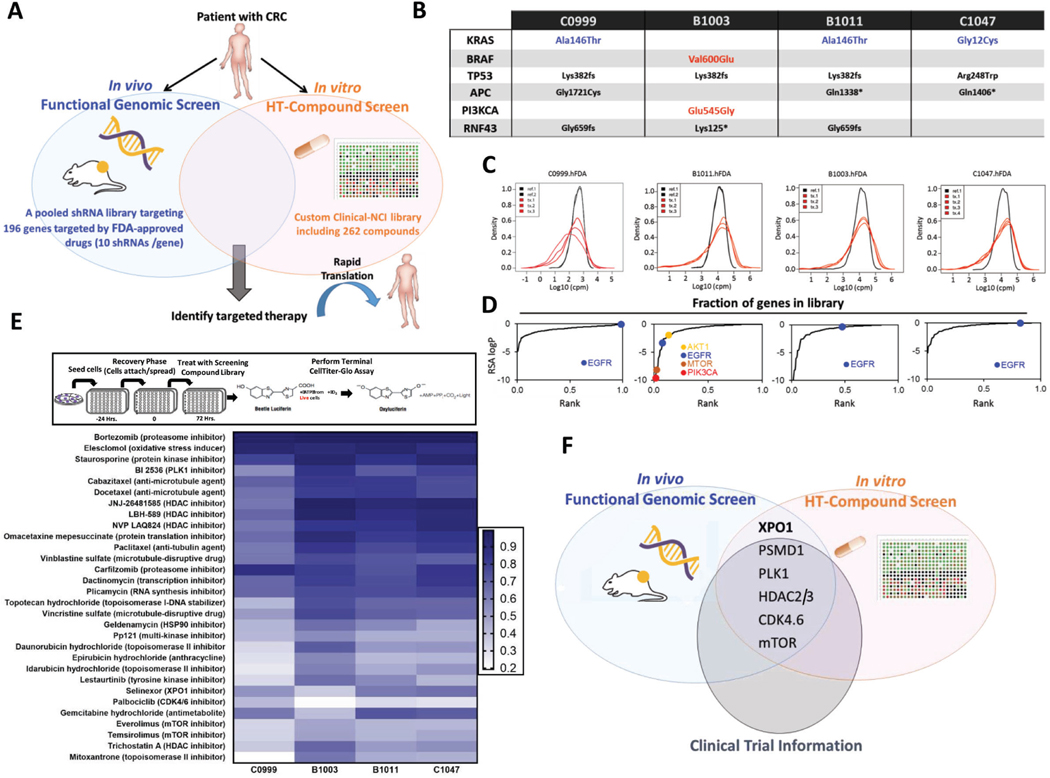
Figure 1.
Integrated genomic and pharmacologic screening using CRC PDX models to identify therapeutic opportunities (A) Schematic of orthogonal screening platform: in vivo shRNA screens in CRC PDXs using a pooled genetic library targeting products of 196 FDA-approved or under clinical investigation genes was combined with in vitro high-throughput compound screens using a Custom Clinical NCI-library including 262 compounds. (B) Genetic landscape of 4 CRC PDXs in the in vitro and in vivo screening pipeline. Three out of four models displayed KRAS mutations (C0999, B1011 and C1047); one of the models harbored BRAF/PIK3CA mutation (B1003). (C) Density distribution of barcodes (shRNA) for transduced PDX cells (References) and three in vivo tumor replicates (Tx 1, 2 and 3) from 4 CRC PDXs infected with the FDAome shRNA lentiviral library. (D) Fraction of scoring genes in the library. Gene-rank analysis highlighting behavior of EGFR, AKT1, mTOR and PIK3CA hits in the FDAome in vivo screens executed in 4 independent CRC PDX models: C0999, B1003, B1011 and C1047 (RSA = redundant shRNA activity, logP). (E) Schematic of the high-throughput drug screen workflow and heatmap of the 30 most potent compounds and their AUCs for the 4 PDX models’ responses to drug exposure in vitro. Results were classified into 4 groups calculated by the extension of the area under the curve (AUCn; Class 1: AUCn≥0.7; Class2: AUCn≥0.4; Class3: AUCn>0.1; Class4: AUCn<0.1). (F) Topscoring genes and corresponding compounds were prioritized for investigation by integrating the orthogonal screening results with currently available clinical trial information in CRC.