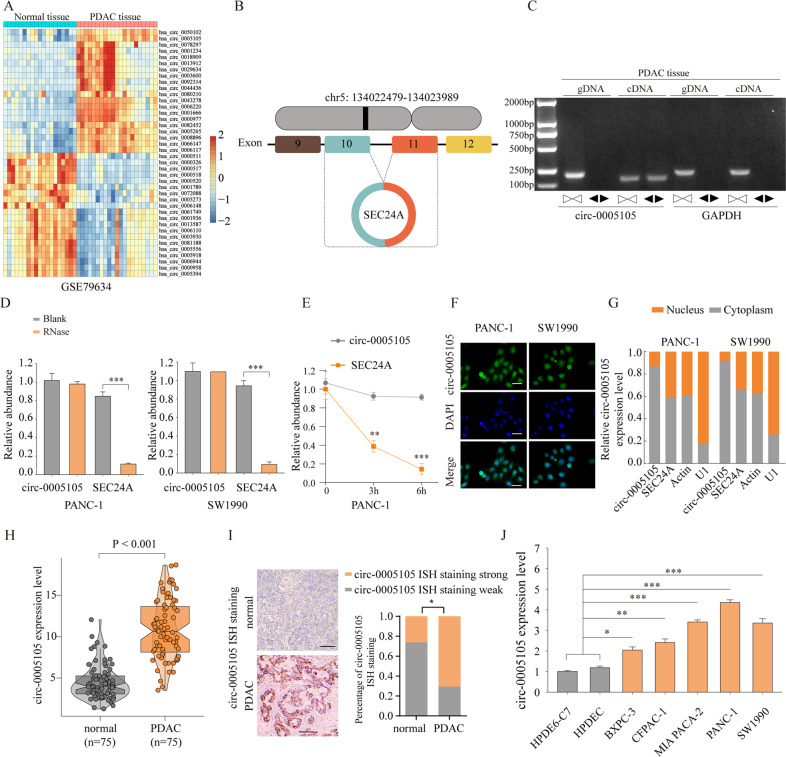
Fig. 1. CircRNA circ-0005105 is overexpressed in PDAC tissues.
A Heat maps of differently expressed circRNAs in PDAC tissue compared with adjacent normal tissue (GSE79634). Red indicates a higher fold-change and blue indicates a lower fold change. B circ-0005105 is produced at the SEC24A gene locus containing exon 9-12. C. RT-PCR assay with divergent or convergent primers indicated the existence of circ-0005105 in PDAC tissue. GAPDH was used as a negative control. qRT-PCR analysis of the expression of circ-0005105 after RNase R treatment (D) and Actinomycin D (E). The subcellular localization of circ-0005105 in PANC-1 and SW1990 cells were analyzed through fluorescence in situ hybridization (FISH) (F) and nuclear and cytoplasmic separation assay; Scale bar = 40 μm (G). The expression of circ-0005105 in 75 pairs of PDAC (Tumor) and adjacent normal tissues (Normal) was determined by qRT-PCR (H) and in situ hybridization (ISH); Scale bar = 400 μm (I). qPCR analysis of circ-0005105 expression in pancreatic epithelial cells (HPDE6-C7 and HPDEC) and pancreatic cancer cells (BXPC-3, CFPAC-1, MIA PACA-2, PANC-1, and SW1990). The results are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.01.