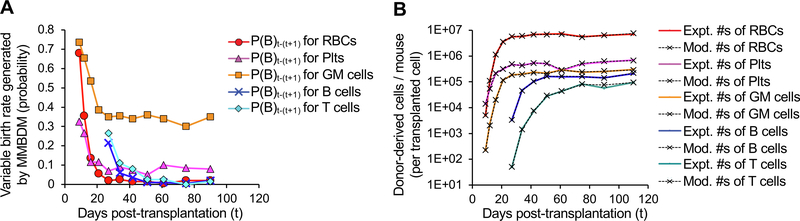
Fig. 3.
Markov-based modeling of birth–death probabilities and the effects on reconstitution profiles. Example of data generated by Modified Markov Birth-Death Model (MMBDM) for both the estimated birth probabilities (A) and the corresponding model fitted cell number values (B). (A) The MMBDM was used to estimate the birth rate (probabilities) between two experimentally assessed time points by taking into account the assumed death rates (probabilities) from literature for each of the 5 mature lineages and the number of cells detected experimentally at both the time points. The birth probability (plotted here) is calculated depending on the closest fit of the model generated numbers to the actual number of cells present at the two time points. The birth rate (in days) can then be estimated between successive time points using the formula: Birth rate in days between time t and t + 1 = Loge(2)/P(B)t-(t+1) where P(B)t–(t+1) = Birth rate probability plotted at time t, for time between t and t + 1. (B) Experimentally observed donor cell derived mature cells per mouse compared to the numbers generated by MMBDM. The overlapping curves (colored [experimental] line with the corresponding black dashed [modeled] line for each population) shows that we achieved high accuracy using the MMBDM to estimate the number of cells present at the various time points. Expt. #s = Experimentally derived numbers at time t; Mod. #s = MMBDM-based numbers at time t.