Abstract
Phytochelatins (PCs) play important roles in the detoxification of and tolerance to heavy metals in plants. The synthesis of PCs is catalyzed by phytochelatin synthase (PCS), which is activated by heavy metal ions. In this study, we isolated a PCS gene, BnPCS1, from the bast fiber crop ramie (Boehmeria nivea) using the RACE (rapid amplification of cDNA ends) method. The full-length BnPCS1 cDNA is 1,949 bp in length with a 1,518 bp open reading frame (ORF) that encodes a 505 amino acid protein. The deduced BnPCS1 protein has a conserved N-terminus containing the catalytic triad Cys58, His164, Asp182, and a flexible C-terminal region containing a C371C372QETC376VKC379 motif. The BnPCS1 promoter region contains several cis-acting elements involved in phytohormone or abiotic stress responses. Subcellular localization analysis indicates that the BnPCS1-GFP protein localizes to the nucleus and the cytoplasm. Real-time PCR assays show that the expression of BnPCS1 is significantly induced by cadmium (Cd) and the plant hormone abscisic acid (ABA). Overexpression lines of BnPCS1 exhibited better root growth and fresh weight, lower level of MDA and H2O2, and higher Cd accumulation and translocation factor compared to the WT under Cd stress. Taken together, these results could provide new gene resources for phytoremediation of Cd-contaminated soils.
Keywords: Boehmeria nivea, phytochelatin synthase, cadmium, accumulation, translocation
Highlights
-
-
The expression of BnPCS1 is significantly induced by Cd and ABA.
-
-
Overexpression of BnPCS1 confers enhanced Cd tolerance, accumulation and translocation in Arabidopsis.
-
-
Our research results could provide new gene resources for phytoremediation.
Introduction
Heavy metal pollution has been recognized one of the most important environmental issues worldwide. When the concentrations of heavy metal ions in soils reach a certain level, plants growing in these soils will exhibit various symptoms of poisoning, including growth retardation, stunting, chlorosis, and finally growth cessation. However, plants have developed diverse adaptive mechanisms during the course of evolution, and some plants display stronger tolerance to the effects of heavy metals (Emamverdian et al., 2015). Phytochelatins (PCs) are small thiolate peptides produced immediately after exposure to heavy metal ions such as Cd2+, As2+, Pb2+, Cu2+, and Zn2+, that exist widely in certain bacteria, algae, fungi, and almost all plant species. PCs have strong chelation activity and play important roles in the accumulation and detoxication of heavy metal ions in plants by chelating these toxic ions to form complexes. The complexes are then sequestered to specific organelles (mainly to the vacuoles) and fixed by a process known as compartmentalization so that the concentration of toxic metal ions will be reduced to levels that plants can tolerate (Kim et al., 2019).
PCs have the general structure (γ-Glu-Cys)n-Gly (n = 2–11) and are synthesized from glutathione (GSH, γ-Glu-Cys-Gly), catalyzed by phytochelatin synthase (PCS, EC 2.3.2.15) (Ramos et al., 2007). PCS belongs to the papain superfamily; it is actually a dipeptidyl transferase in which the catalytic reaction process is similar to that of cysteine protease. The synthesis of PCs can be divided into two stages: in the first stage, the γ-Glu-Cys unit from GSH is cleaved and transferred to the free enzyme to generate a kind of acyl-enzyme intermediate. In the second stage, the γ-Glu-Cys unit is transferred from the intermediate to another substrate molecule which can be GSH or an oligomeric phytochelatin peptide (PCn) to form PCn+1. The cycle is then repeated until the final products (γ-Glu-Cys)2–11-Gly, are produced (Vatamaniuk et al., 2004). The reactions are shown below (Rigouin et al., 2013):
Stage 1: γ−Glu−Cys−Gly+PCS→γ−Glu−Cys−PCS+Gly
Stage 2: γ−Glu−Cys−PCS+(γ−Glu−Cys)nGly→PCS+(γ−Glu−Cys)n+1−Gly
In the fission yeast (Schizosaccharomyces pombe) and various plant species, the amino acid sequences of most eukaryotic PCSs consist of conserved N-terminal and variable C-terminal domains. The N-terminal domains from most organisms share high levels of sequence homology, and are suggested to have catalytic activity (Hayashi et al., 2020). There are several conserved Cys residues in the N-terminal domains, among which the Cys57 residue is present in almost all known PCS proteins, and it may be related to the catalytic activity of PCS enzymes. The C-terminal domain is considered to function in regulating metal ion activity, and contains pairs of Cys and Glu residues. In the absence of heavy metals, the N-terminal domains have no enzymatic activity. When heavy metals are detected, the C-terminal domain forms a special structure with the heavy metal ions that can initiate catalytic activity in the N-terminal domain. The ability of different metal ions to induce catalytic activity in PCS enzymes varies, and Cd2+ has been shown to be the most efficient metal catalyst (Filiz et al., 2019).
A PCS enzyme with biological activity was first isolated from Silene cucubalus in 1989 (Grill et al., 1989), followed by similar reports from Arabidopsis thaliana (Vatamaniuk et al., 1999), Pteris vittata (Dong, 2005), Sesbania rostrata (Li et al., 2009), Triticum aestivum (Couselo et al., 2010), tall fescue (Zhao et al., 2014) Oryza sativa (Das et al., 2017), and Ipomoea pes-caprae (Su et al., 2020). AtPCS1-deficient Arabidopsis plants are highly sensitive to Cd, while overexpression of AtPCS1 changed the Cd tolerance and the ability of plants to accumulate Cd (Ha et al., 1999; Pomponi et al., 2006). Tobacco plants expressing NtPCS1 showed an increased tolerance to arsenic (As) and Cd, but changes in the accumulation of As and Cd were not observed (Lee and Hwang, 2015). Heterologous expression of the CdPCS1 gene from Ceratophyllum demersum in Arabidopsis and Escherichia coli enhanced the accumulation of heavy metals (Shukla et al., 2012).
Boehmeria nivea (L.) Gaudich., commonly known as ramie, is a perennial herb in the nettle family (the Urticaceae) that is native to eastern Asia. Ramie is also known as “China grass,” and the fibers are widely used in the textile industry to make fabric. Plants of B. nivea have well-developed root systems, fast growth, high reproducibility, strong resistance to stress and disease, and produce a large amount of biomass, characteristics that can quickly make up for the shortages of other known heavy metal hyperaccumulators. In addition, ramie is used as a raw industrial material that will not enter the food chain and can produce economic benefits. Previous studies have shown that ramie has strong heavy metal tolerance and the ability to accumulate several heavy metals from soil (She et al., 2011; Shukla et al., 2012). However, the mechanisms that determine Cd tolerance in ramie are unclear. In our previous studies on the transcriptome profiles of Cd-responsive genes in ramie, unigene6921 was found to be significantly up-regulated in the Cd treatment group, and this gene was annotated as a phytochelatin synthase gene (She et al., 2015). In this paper, we isolated the full-length cDNA of the PCS gene unigene6921 using the RACE method (rapid amplification of cDNA ends). The gene is designated BnPCS1, and our study focused on gene promoter analysis, subcellular localization of the BnPCS1 protein, gene expression characteristics, and the possible functions of BnPCS1 in the response to Cd stress in B. nivea.
Materials and Methods
Plant Growth Conditions and Hormone and Cd Treatments
The B. nivea cultivar “Zhongzhu No. 1” used as plant material in this study was obtained from the Yunyuan farm of Hunan Agricultural University, Changsha, China. The ramie seedlings were grown hydroponically in half-strength Hoagland nutrient solution for 21 days. For the hormone treatments, the seedlings were sprayed with 100 μM abscisic acid (ABA) or 1 mM salicylic acid (SA). For the Cd treatment, seedlings were transferred to half-strength Hoagland solution containing 200 μM Cd. Roots, stems, and leaves were harvested after 0, 2, 4, 6, 12, and 24 h of ABA or SA treatment. Leaves were harvested after 0, 3, 6, 9, 12, 24, and 48 h of Cd treatment. The roots and shoots were frozen immediately in liquid nitrogen and stored at −80°C. For each time point there were three biological replicates.
Isolation of RNA and Genomic DNA and First-Strand cDNA Synthesis
Total RNA was extracted from plant tissues using the RNAprep pure Plant Kit (Tiangen, Beijing, China). For first-strand cDNA synthesis, ∼1.0 μg of total RNA was used as the template in the PrimeScript RT reagent Kit with gDNA Eraser Kit (Takara, Beijing, China) following the manufacturer’s instructions. Genomic DNA was extracted from B. nivea tissues using the Rapid Plant Genomic DNA Isolation Kit (Shanghai Shenggong Co., Shanghai, China) as directed by the manufacturer.
Isolation of the Full-Length BnPCS1 cDNA
To obtain the full-length cDNA sequence of BnPCS1, the 5′RACE primers PCS1-5FO/5FI and the 3′RACE primers PCS1-3FO/3FI (Supplementary Table 1) were designed based on the sequence of unigene6921 from the ramie transcriptome using Prime Premier 5.0 software. The 5′RACE and 3′RACE reactions were performed using the SMARTer RACE5′/3′ Kit (Clontech, United States) following the manufacturer’s protocol. The products from 5′RACE and 3′RACE were examined on 1.5% agarose gels, purified with the TaKaRa MiniBEST Agarose Gel DNA Extraction Kit (Takara, Beijing, China), and cloned into the pMD18-T vector (Takara, Beijing, China) prior to DNA sequencing. The sequences from 5′ RACE, 3′RACE, and the known gene fragment were then assembled using DNAMAN 8.0 software. To verify the accuracy of the assembled sequence, a pair of primers PCS1-F/R (Supplementary Table 1) was used to amply the full-length sequence which was then sequenced at Shanghai Shenggong Company (Shanghai, China). The nucleotide sequence of BnPCS1 was submitted to the National Center for Biotechnology Information (NCBI) Genbank database under Accession No. KF717368.
Isolation of the BnPCS1 Gene Promoter
Genomic DNA extracted from ramie cultivar “Zhongzhu No. 1” was used as the template to amplify the 2,057 bp of genomic DNA sequence upstream from the BnPCS1 initiation codon using the primer pair PCS1-PF/PR (Supplementary Table 1). The PCR amplification proceeded for 32 cycles of denaturation at 98°C for 10 s, primer annealing at 55°C for 15 s, and extension at 72°C for 30 s using PrimeSTAR Max DNA Polymerase (Takara, Beijing, China). The PCR product was examined by agarose gel electrophoresis, purified, and sequenced. The cis-acting regulatory elements in the BnPCS1 promoter region were predicted by searching the PlantCARE database1.
Sequence Analysis
The BnPCS1 ORF was analyzed and translated with the ORF Finder2. The physicochemical properties of the amino acid sequences deduced from BnPCS1 were predicted with the ProtParam tool on the ExPASy server3. The sequence comparison was performed using the BLASTP tool4. The conserved domains in the BnPCS1protein were predicted with the Conserved Domain Search Service (CD Search)5. Multiple sequence alignment was performed with DNAMAN 8.0 software. A phylogenetic tree showing the evolutionary relationships between BnPCS1 and predicted PCS proteins from other plant species was constructed with MEGA 5.0 software using the neighbor-joining (NJ) method based on Kimura’s 2-parameter distance and 1,000 bootstrap replicates to estimate the confidence of the individual branches. The protein subcellular localization was predicted withWoLF PSORT6.
Subcellular Localization of BnPCS1 Protein
A pair of specific primers, PCS1-XbaI-F and PCS1-SmaI-R (Supplementary Table 1), was used to amplify the BnPCS1 ORF. The PCR product was digested with the restriction enzymes XbaI and SmaI and cloned into the expression vector pAN580, so that the BnPCS1 protein is fused to the green fluorescent protein (GFP) reporter gene toconstruct plasmid 35S-BnPCS1-GFP. The empty vector 35S::GFP was used as the control while the 35S::OsGhd7-CFP plasmid was used to produce the nuclear marker. The constructs were separately introduced into Arabidopsis protoplasts. Protoplast isolation and transfection were performed according to the method described by Yoo et al. (2007). Briefly, mesophyll protoplasts were isolated from the rosette leaves collected from 3 to 4-weeks-old Col wild type Arabidopsis thaliana plants. Then, the nuclear marker construct 35S::OsGhd7-CFP was co-transformed with 35S-BnPCS1-GFP constructor 35S::GFP construct into Arabidopsis protoplasts. The fluorescence signals of GFP and CFP (cyan fluorescentprotein) were observed with a confocal microscope imaging system (Olympus FV1200) after the protoplasts were incubated at room temperature for 20–22 h in the dark.
Real-Time Quantitative RT-PCR Analysis
Real-time quantitative RT-PCR (qRT-PCR) was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems). The ramie actin gene (DQ665832) was amplified along with BnPCS1 as a reference gene to normalize gene expression. For tissue-specific expression analysis, total RNA was extracted from the roots, stems, stem tips, and leaves of ramie seedlings. First-strand cDNA synthesis was carried out using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Beijing, China) following the manufacturer’s instructions. For gene expression analysis of plants treated with Cd, cDNA was synthesized from total RNA extracted from roots, stems, and leaves of seedlings treated with Cd solution at different time points. The nucleotide sequences of the primers used to amplify BnPCS1 (PCS1-qF/R) and the actin gene (BnActin-F/R) are given in Supplementary Table 1. Quantification of the relative transcript levels was performed using the comparative 2–ΔΔCt method (Livak and Schmittgen, 2001). The qRT-PCR assays were conducted with three biological replicates and three independent technical replicates for each sample.
Over-Expression of BnPCS1 in Arabidopsis
To over-express BnPCS1 in Arabidopsis, the complete BnPCS1 ORF was amplified by PCR with a pair of gene-specific primers with added 5’ restriction enzyme sites (PCS1-SmaI-F and PCS1-BamHI-R). The PCR amplification proceeded for 32 cycles and consisted of denaturation at 98°C for 10 s, primer annealing at 60°C for 15 s, and extension at 72°C for 30 s. After double-digestion with restriction enzymes SmaI and BamHI, the PCR-amplified product was subcloned into the plant expression vector pBI121 to make the construct named pBI121-BnPCS1. The recombinant plasmid was introduced into the Agrobacterium tumefaciens strain EHA105 using the freeze-thaw method, and the construct was then transformed into Arabidopsis Col-0 plants using the floral-dip method (Clough and Bent, 1998; Zhang et al., 2006). Briefly, Agrobacterium cultures were grown on a platform shaker (220 r.p.m.) at 28°C to stationary phase in sterilized LB medium (containing 10 g tryptone, 5 g NaCl, 5 g yeast extract per liter water) carrying added kanamycin (30 mg⋅L–1). Cultures were typically started from a 1:100 dilution of smaller overnight cultures and grown for roughly 18–24 h. Cells were harvested by centrifugation at 5,500 × g for 20 min and resuspended in infiltration medium (containing 5% sucrose, 0.05 % Silwet L-77, 2.15 g⋅L–1 MSsalts) to a final OD600 = 0.8. Then, the medium was added to a beaker, plants were inverted into this medium such that all above-ground parts were submerged, and plants were removed after 3–5 s of gentle agitation. Plants were removed from the beaker, domed to remain humidity and covered with black cloth. The cloth was removed the next day and plants were regularly watered until seeds maturation. Seeds (T3 generation) of the homozygous transgenic lines were screened on 1/2 strength MS medium containing 30 mg⋅L–1 kanamycin. The kanamycin resistant lines were further verified for the presence of the BnPCS1 gene by PCR and real-time RT-PCR. The primers PCS1-35S-F and PCS1-SP-R used for PCR verification are given in Supplementary Table 1.
Cd Stress Assay
Phenotypic Analysis
To test the effect of Cd stress on the growth of transgenic Arabidopsis seedlings, seeds from the transgenic lines (T3 generation) and wild-type (WT) plants were cultured on half-strength solid Murashige and Skoog medium (per liter add 100 ml 10 × macronutrients, 10 ml 100 × micronutrients, 5 ml 1% Fe-EDTA, 30 g sucrose, pH 5.6–5.8, with 1 M KOH, 7 g agar) (Murashige and Skoog, 1962) containing Cd (0, 100, or 150 μM). The fresh weights and root lengths of transgenic and wild-type Arabidopsis seedlings were measured at 14 days, and the plants were photographed. WT seedlings were used as the control. There were three experimental replicates.
Determination the Cd Content in Plants
The transgenic and wild-type Arabidopsis were planted in plastics pots with unpolluted vermiculite and irrigated with half-strength Hoagland nutrient solution containing 2.5 mM Ca(NO3)2, 2.5 mM KNO3, 1 mM MgSO4, 1 mM KH2PO4, and 23.1 μM H3BO3, 4.6 μM MnCl2, 0.19 μM CuSO4, 1.2 μM ZnSO4, 0.12 μM Na2MoO4 and 45 μM Fe(III)-EDTA at pH 6.0 (Terry, 1980) for 28 days. Then, CdCl2 salt was added to the half-strength Hoagland nutrient solution to the final concentration of 50 μM, and the plants were watered with the Cd solution one time. After 14 days of cultivation, the plants were harvested, washed with tap water and rinsed with deionized water three times. Then, the shoots and roots were separated and dried to a constant weight (65°C for 72 h). After that, Cd in the samples was measured by flame AAS with HNO3-HClO4 digestion. There were three experimental replicates.
Translocation factor (TF) is calculated from the ratio of cadmium’s presence in the plant shoots compared to that in roots using the equation:
Determining the H2O2 Content
Twenty-one-day-old seedlings from the transgenic lines (T3 generation) and WT cultured in soil without Cd were irrigated with 0, 100, or 150 μM Cd solution and grown for another 2 days. The H2O2 content was determined using the method described by DeLong et al. (2002). Briefly, a 1-gram sample of leaf tissue was ground into a powder and extracted with 80 percent ethanol. The homogenate was then centrifuged at 10,000 × g for 10 min, and 0.1 mL of the supernatant was added to 1 mL working solution (containing 100 μM xylenol orange, 250 μM ammonium ferrous sulfate hexahydrate, 90 percent methanol, and 25 mM H2SO4) and incubated for 30 min at 30°C. The absorbance of the reaction mixtures was measured at 560 nm with a UV-visible spectrophotometer (UV-1800, Shimadzu, Japan). The standard curve was established using H2O2 concentrations ranging from 0 to 100 μM to calculate the content of H2O2 in the unknown samples.
Determining the MDA Content
Twenty-one-day-old seedlings from the transgenic lines (T3 generation) and WT plants cultured in the soil without Cd were irrigated with 0, 100, or 150 μM Cd solution and further grown for 14 days. The content of MDA was determined according to the method described by Hodges et al. (1999). Leaf tissue samples (0.5 g) were ground to powder and extracted with 10 mL of 20 percent trichloroacetic acid (TCA). The homogenates were centrifuged at 10,000 × g for 10 min, and 2 mL of supernatant from each extract was added to 2 mL of 0.6 percent thiobarbituric acid (TBA) solution containing 10 percent TCA and incubated at 100°C for 15 min. The reaction mixtures were then rapidly cooled in an ice bath. The absorbance of each supernatant was measured at 450, 532, and 600 nm using a UV-Visible spectrophotometer (UV-1800, Shimadzu, Japan). The MDA content was calculated as 6.45 × (A532 - A600) - 0.56 × A450.
Statistical Analysis
Statistical analyses were conducted using SPSS version 17.0 and Microsoft Excel 2013 software. All data were expressed as the mean of three biological replicates ± standard deviation (SD). Comparisons between different groups were tested by one-way ANOVA followed by Student’s t-test. A p-value < 0.05 was considered to indicate a significant difference.
Results
Isolation and Characterization of BnPCS1
The 1,949 bp full-length cDNA for the BnPCS1 gene was isolated using 3′-RACE, 5′-RACE, and RT-PCR methods. DNA sequence analysis showed that the cDNA contains a 1,518 bp open reading frame (ORF) that is predicted to encode a 505 amino acid protein (Supplementary Figure 1). The physicochemical properties of the predicted BnPCS1 protein were analyzed using the ExPASy database, which showed that the BnPCS1 protein has a predicted molecular mass of 56.02 kDa and an isoelectric point of 7.01. The secondary structure of the BnPCS1 protein was predicted using the NPS@(Network Protein Sequence Analysis) server, which indicated that the BnPCS1 protein consists of α-helices (49.70%), extended strands (11.49%), and random coils (38.81%). The protein subcellular localization predicted using WoLFPSORT showed that BnPCS1 protein is mainly located in both nucleus and cytoplasm.
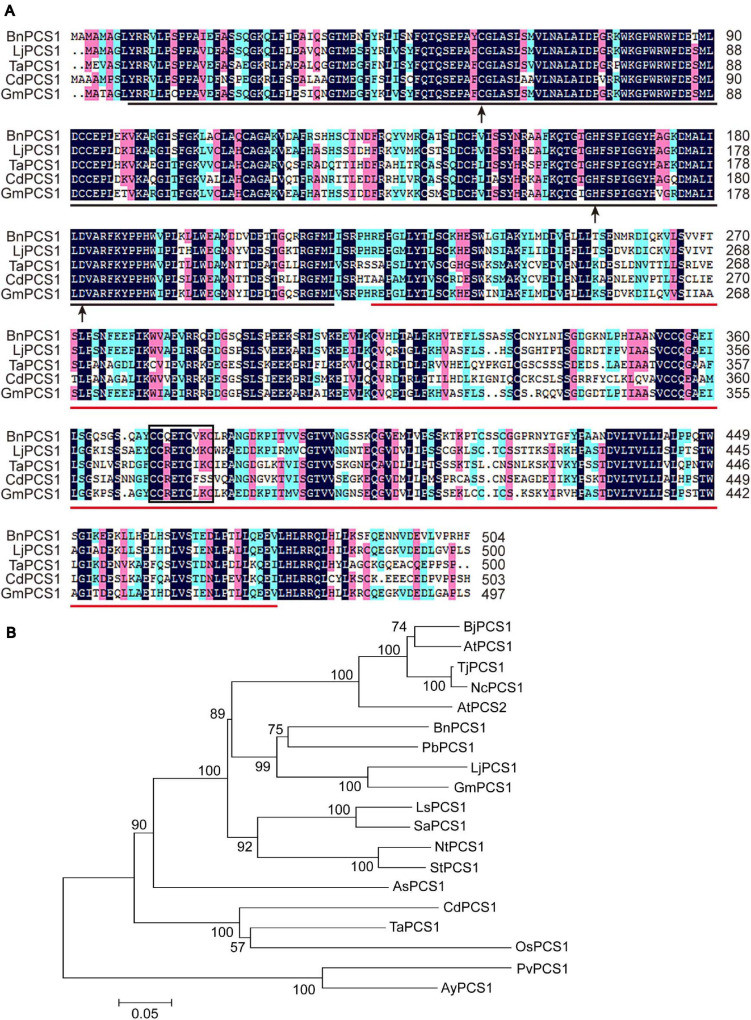
A conserved domain search of the NCBI database showed that the deduced amino acid sequence of BnPCS1 contains two domains: a phytochetatin domain at amino acids 9–217 and a phytochelatin_C domain at amino acids 223–478. There are three active sites in the phytochetatin domain; (1) the cysteine residue at amino acid residue 58, (2) the histidine residue atamino acid residue 164, and (3) the aspartic acid residue at amino acid residue 182. A C371C372QETC376VKC379 motif in the PCS serves as a sensor for heavy metal ions and was found within the phytochelatin_C domain (Figure 1A). BLASTp searches and multiple sequence alignments revealed that the BnPCS1 protein shares a high degree of sequence similarity with the reported phytochelatin synthases from other plant species such as Lotus japonicus (GenBank accession number: AAT80342.1; 73%), Triticum aestivum (AAD50592.1; 61%), Cynodon dactylon (AAO13810.2; 57%), and Glycine max (AAL78384.1; 73%). In addition, the N-terminal part of the PCS proteins is more conserved than the C-terminal part (Figure 1A).
FIGURE 1.
Multiple sequence alignment and phylogenetic analysis of BnPCS1 and PCS proteins from other plant species. (A) Alignment of BnPCS1 and its orthologous proteins from various plant species constructed using DNAMAN 8.0 software. The shaded regions indicate the conserved amino acid residues (those shown in black are fully conserved; the amino acids highlighted in pink and light green are similar). The region underlined in black is the phytochetatin domain, and the phytochelatin_C domain is underlined in red. The three active sites are indicated by black arrows. The C371C372QETC376VKC379 motif is enclosed in a black box. (B) Phylogenetic tree showing the evolutionary relationships between BnPCS1 and 18 other PCS proteins from various plant species constructed using the neighbor-joining (NJ) method in MEGA 5.0. The scale bar represents 0.05 amino acid substitutions per site. GenBank accession numbers of the proteins are as follows: AtPCS1 (AAD16046.1) and AtPCS2 (AAK94671.1) from Arabidopsis thaliana, BjPCS1 (CAC37692.1) from Brassica juncea, TjPCS1 (BAB93119.1) from Thlaspi japonicum, NcPCS1 (BAB93120.1) from Noccaea caerulescens, PbPCS1 (AEY68568.1) from Pyrus betulifolia, LjPCS1 (AAT80342.1) from Lotus japonicus, GmPCS1 (AAL78384.1) from Glycine max, LsPCS1 (AAU93349.1) from Lactuca sativa, SaPCS1 (ACU44656.1) from Sonchus arvensis, NtPCS1 (AAO74500.1) from Nicotiana tabacum, StPCS1 (CAD68110.1) from Solanum tuberosum, AsPCS1 (AAO13809.1) from Allium sativum, CdPCS1 (AAO13810.2) from Cynodon dactylon, TaPCS1 (AAD50592.1) from Triticum aestivum, OsPCS1 (AAO13349.2) from Oryza sativa, and PvPCS1 (AAT11885.1) and AyPCS1 (BAB64932.1) from the ferns Pteris vittata and Athyrium yokoscense, respectively.
To analyze the evolutionary relationships of PCS proteins, the sequences of BnPCS1 and 18 previously reported PCS proteins from various plant species were downloaded from GenBank and used to construct a phylogenetic tree using the neighbor-joining method as implemented in MEGA 5.0 software (Tamura et al., 2007). The phylogenetic tree showed that BnPCS1 has the closest evolutionary relationship with PbPCS1, LjPCS1, and GmPCS1, which come from dicotyledonous plants in the order Rosales. BnPCS1 is more distantly related to CdPCS1, TaPCS1, OsPCS1, PvPCS1, and AyPCS1, which belong to two clades of PCS1 proteins from monocotyledonous plants (Figure 1B).
Isolation of the BnPCS1 Promoter
A pair of primers, PCS1-PF and PCS1-PR (Supplementary Table 1), were designed to isolate a 1,928 bp DNA fragment containing the BnPCS1 promoter (Supplementary Figure 2). Analysis ofthe cis-acting elements in the promoter sequence using searches of the PlantCARE database indicated that the promoter contains not only the basic promoter elements such as the CAAT-box and TATA-box, but also some cis-acting elements associted with stress responses, such as an anaerobic induction element (ARE), a drought responsive element (DRE1), a stress responsive element (STRE), and a wound responsive element (WUN-motif). Also present were some hormone response elements, such as an abscisic acid responsive element (ABRE), an MeJA responsive element (CGTCA-motif), a salicylic acid responsiveelement (TCA-element), and an auxin responsive element (TGA-element) (Table 1). These results suggest that BnPCS1 may participate in responses to abiotic and biotic stresses.
TABLE 1.
Putative cis-acting regulatory elements identified inthe promoter region sequence of BnPCS1 using thePlantCARE database.
| Cis element | Position | Sequence | Function of site |
| ABRE | +717 | ACGTG | Cis-acting element involved in the abscisic acid responsiveness |
| ARE | +235 | AAACCA | Cis-acting regulatory element essential for the anaerobic induction |
| CAAT-box | +122, -220, +316, +440, +615, +861, +938, +1018, +1131, -1179, +1253, +1360, -1409, +1787 | CAAT, CAAAT, CCCAATTT, CCAAT | Common cis-acting element in promoter and enhancer regions |
| CGTCA-motif | -427 | CGTCA | Cis-acting regulatory element involved in the MeJA-responsiveness |
| DRE1 | -1725 | ACCGAGA | Cis-acting regulatory element involved in the drought responsiveness |
| G-box | +47, +716 | CACGAC, TACGTG | Cis-acting regulatory element involved in light responsiveness |
| Gap-box | +440 | CAAATGAAAA | Part of a light responsive element |
| MRE | +172 | AACCTAA | MYB binding site involved in light responsiveness |
| STRE | +594, +1150, -1122 | AGGGG | Cis-acting regulatory element involved in stress responsiveness |
| TATA-box | -241, +271, -290, -323, -325, +342, -347, +355, -481, +484, -509, -525, -568, -623, +627, +702, +706, +941, -976, -1025, -1030, +1077, -1090, -1210, -1220, -1241, -1270, -1413, -1599, -1623, -1627 | TACAAAA, TATTTAAA, TACATAAA, TATAAAT, TATATAA, TATAA,TATA, TATATA, TATAT, TATAAAA | Core promoter element around -30 of transcription start |
| TCA-element | -603, +1008 | CCATCTTTTT | Cis-acting element involved in salicylic acid responsiveness |
| TGA-element | -1501, -1748 | AACGAC | Auxin-responsive element |
| WUN-motif | -1318, -1351, -1319 | AAATTACTA, AAATTACT | Cis-acting regulatory element involved in the wound responsiveness |
| Circadian | +778 | CAAAGATATC | Cis-acting regulatory element involved in circadian control |
Subcellular Localization of the BnPCS1 Protein
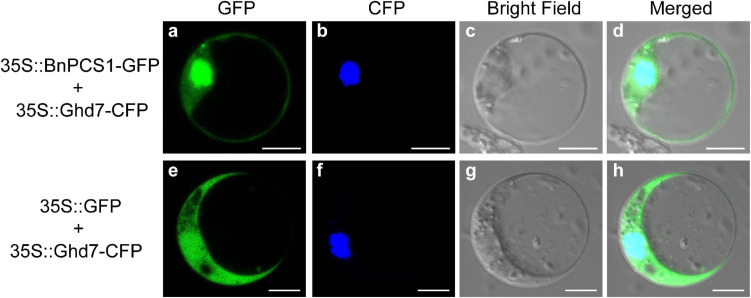
The protein subcellular localization predicted using WoLF PSORT indicated that BnPCS1 protein is mainly localized in both nucleus and cytoplasm. To verify this prediction, the recombinant plasmid 35S::BnPCS1-GFP, which contains an enhanced GFP gene fused to BnPCS1 in pAN580, was transfected into Arabidopsis protoplasts, the empty 35S::GFP vector wasused as the control. Microscopic observation showed that the green fluorescence in cells transfected with 35S::BnPCS1-GFP were observed both in the nucleus and the cytoplasm along with the cyan fluorescent signal of nuclear marker OsGhd7-CFP (Figures 2a–d). In contrast, green fluorescence was detected throughout the cells expressing the GFP (Figures 2e–h) gene alone. The above results indicated that BnPCS1 protein is localizedin both the nucleus and cytoplasm of plant cells.
FIGURE 2.
Subcellular localization of BnPCS1. Recombinant plasmid 35S::BnPCS1-GFP and the empty vector 35S::GFP were transfected separately into Arabidopsis protoplasts with nuclear marker 35S::OsGhd7-CFP. The CFP and GFP fluorescence signals were observed using a laser confocal microscope. (a) 35S::BnPCS1-GFP; (b) 35S::OsGhd7-CFP; (c) bright field; (d) overlap images of (a–c); (e) 35S::GFP; (f) 35S::OsGhd7-CFP; (g) bright field; (h) overlap images of (e–g).
Gene Expression Analysis of BnPCS1 in B. nivea
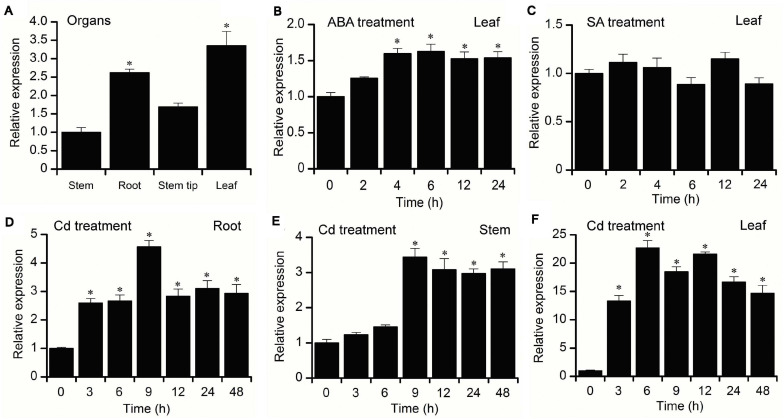
To examine the tissue-specific expression characteristics of BnPCS1, the transcriptional levels of BnPCS1 in the root, stems, stem tip, and leaves of 21-day-old B. nivea seedlings were analyzed using quantitative real-time PCR (qRT-PCR). The results showed that the highest expression level of BnPCS1 was in leaves, and the lowest expression level was in the stems (Figure 3A). For the hormone treatments, the 21-day-old B. nivea seedlings were sprayed with 100 μM abscisic acid (ABA) or 1 mM salicylic acid (SA), respectively. Expressions of BnPCS1 in B. nivea leaves after treatment with ABA or SA for 0, 2, 4, 6, 12 or 24 h were assayed by qRT-PCR. We found that the relative expression of BnPCS1 is rapidly induced by ABA and reached its highest level after 6 h, which was 1.6-fold the level in untreated leaves (Figure 3B). However, the expression of BnPCS1 was not significantly induced by SA treatment (Figure 3C). To investigate the effects of Cd treatment on the expression of BnPCS1, the mRNA levels of BnPCS1 in B. nivea were examined by qRT-PCR after treatment with 200 μM Cd for 0, 3, 6, 9, 12, 24, or 48 h. The results indicated that BnPCS1 is significantly up-regulated by Cd treatment in the roots, stems, and leaves of B. nivea seedlings. The highest expression levels of BnPCS1 in the roots, stems, and leaves were 4. 6-, 3. 4-, and 22.7-fold higher than in the untreated samples, respectively (Figures 3D–F).
FIGURE 3.
Expression pattern of BnPCS1. (A) qRT-PCR analysis of BnPCS1 mRNA levels in the roots, stems, stem tips, and leaves of 21-day-old B. nivea seedlings. (B,C) BnPCS1 expression levels in the leaves of B. nivea seedlings treated with 100 μM ABA (B) or 1 mM SA (C) for 0, 2, 4, 6, 12, and 24 h. (D–F): BnPCS1 expression levels in the roots (D),stems (E), and leaves (F) of B. nivea seedlings treated with 200 μM Cd for 0, 3, 6, 9, 12, 24, and 48 h. Data are presented as the means of three biological replicates with SE shown by vertical bars. Asterisks indicate significant differences (p < 0.05) between the treatment groups and the controls.
Overexpression of BnPCS1 Enhanced Cd Tolerance and Accumulation in Transgenic Arabidopsis Plants
To investigate whether BnPCS1 is involved in the response of plants to Cd, the complete coding region of BnPCS1 was cloned and inserted into the binary vector pBI121 to generate the pBI121-BnPCS1 construct in which BnPCS1 was over-expressed under control of the CaMV 35S promoter. The recombinant plasmid in Agrobacterium tumefaciens strain EHA105 was then transformed into Arabidopsis ecotype Columbia (Col-0) using the floral-dip method. Seeds obtained from the T3 generation of transgenic Arabidopsis plants were screened on half-strength MS medium supplemented with 30 mg⋅L–1 kanamycin (Supplementary Figure 3). Of these, five transgenic lines (designated L1, L2, L3, L4, and L5) were found to be positive for BnPCS1 following PCR screening (Supplementary Figure 4). Real-time PCR analysis showed that the BnPCS1-specific mRNA levels in the five transgenic Arabidopsis lines varied, and BnPCS1 expression was not detected in WT plants. The highest mRNA level was found in line L1, followed by line L3, and both lines were selected for additional analysis (Supplementary Figure 5).
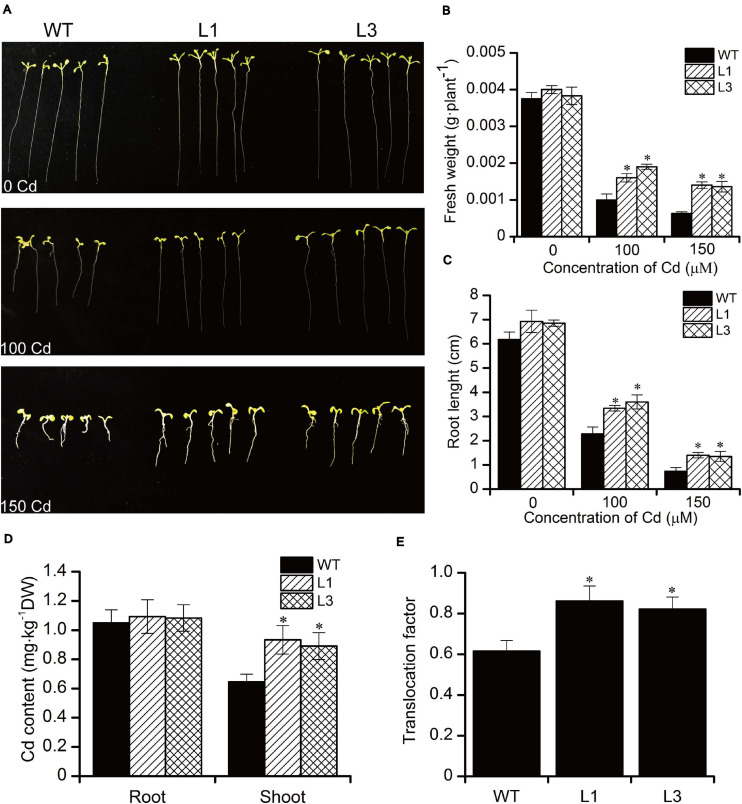
To analyze the effect of Cd stress on the growth of BnPCS1-expressing transgenic Arabidopsis lines (L1 and L3)as compared to WT, seeds of the T3 generation from WT, L1, and L3 were germinated and grown on half-strength MS medium containing three concentrations of Cd (0, 100, and 150 μM). The fresh weights and root lengths of transgenic and wild-type Arabidopsis seedlings were measured at 14 days. There were no significant differences in the phenotypes and growth between the transgenic and WT plants under normal conditions without Cd in the medium. Following Cd treatment, the growth of all plants was inhibited (Figure 4A). However, the fresh weights and root lengths of the transgenic plants were significantly higher than in WT plants. In response to Cd stress, the fresh weight of L1 and L3 line plants were 1.6 and 2.2-fold higher than in WT, respectively; similarly, the roots of L1 and L3 plants were 1.5 and 1.9-fold longer than in WT, respectively (Figures 4B,C). To examine the influence of BnPCS1 on Cd accumulation in Arabidopsis, 28-day-old WT and transgenic Arabidopsis seedlings grown in vermiculite were irrigated with half-strength Hoagland nutrient solution supplement with 50 μM Cd and allowed to grow for 14 days under normal management of water and fertilizer conditions. The Cd content in roots and shoots of WT and transgenic plants were measured. There were no significantly differences of Cd content among WT and two transgenic lines (L1 and L3). However, the two transgenic lines showed significantly higher levels of Cd than WT plants in shoots. Cd content in shoots of L1 and L3 was 0.93 and 0.89 mg⋅kg–1, which was 1.43 and 1.37-fold higher than WT (0.65 mg⋅kg–1), respectively (Figure 4D). We also analyzed the translocation factor (TF) of Cd in WT, L1 and L3. TF is defined as the ratio of metal concentration in plant roots to shoots, which is an important tool used to assess a plant’s potential for phytoremediation purposes. The TF of L1 and L3 was 0.86 and 0.82, which was significantly higher than that of WT (0.61) (Figure 4E). The results indicated that overexpression of BnPCS1 contributes to the translocation of Cd from roots to shootsin transgenic Arabidopsis plants.
FIGURE 4.
Analysis of the response to Cd exposure in WT and BnPCS1 transgenic Arabidopsis plants. (A) Root lengths in Arabidopsis seedlings grown on 0.5× MS medium containing 0, 100, or 150 μM Cd for 14 days. (B,C) Analysis of fresh weights and root lengths of Arabidopsis seedlings grown on 0.5× MS medium containing 0, 100, or 150 μM Cd for 14 days. (D) Cd content in roots and shoots of WT and transgenic Arabidopsis plants grown in vermiculite irrigated with 50 μM Cd for 14 days; (E) Translocation factor (TF) of Cd in WT, L1 and L3. Data are presented as the means of three biological replicates with SE shown by vertical bars. Asterisks indicate significant differences (p < 0.05) between the transgenic lines compared to WT.
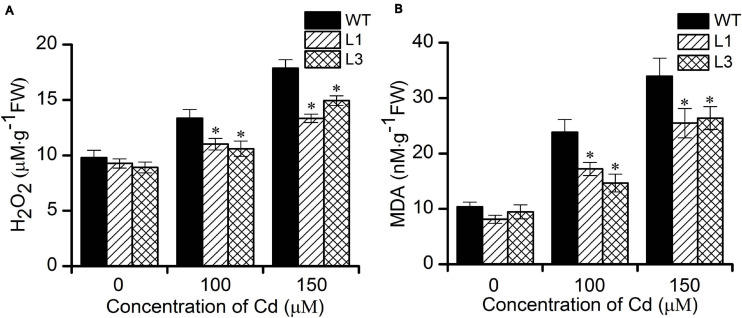
Lipid Peroxidation and Accumulation of H2O2
It is well-known that heavy metal stress induces the production of reactive oxygen species (ROS) such as H2O2 and which can inactivate enzymes and cause cellular damage by degrading proteins and interfering in pathways that are important in cell metabolism (Choudhury et al., 2013). MDA is one of the most well-known secondary products of lipid peroxidation, and it is thus often used as a marker for cell membrane injury (Grotto et al., 2009). To evaluate whether transgenic Arabidopsis have a higher antioxidative capability. We further measured the levels of H2O2 and MDA in WT and transgenic plants. As shown in Figures 5A,B, no significant differences in the H2O2 and MDA contents were found between the WT and transgenic plants. Following treatment with 100 and 150 μM Cd, the concentrations of H2O2 and MDA increased in all plants. However, plants of the transgenic lines L1 and L3 exhibited significantly lower levels of H2O2 and MDA than did the WT plants. These results show that overexpression of BnPCS1 was able to reduce ROS production, which could potentially alleviate the oxidative damage to cells caused by Cd exposure in plants.
FIGURE 5.
Cellular H2O2 and MDA contents in WT and BnPCS1-expressing transgenic Arabidopsis plantsin response to Cd stress. (A) H2O2 contents in WT and transgenic line plants treated with 0, 100,and 150 μM Cd for 2 days. (B) MDA contents in WT and transgenic line plants treated with 0, 100,and 150 μM Cd for 14 days. Data are presented as the means of three biological replicates with SE shown by vertical bars. Asterisks indicate significant differences (p < 0.05) in Cd-treated plants compared to WT.
Discussion
Previous studies have shown that the PC-dependent pathway is a key mechanism for resistance to heavy metals in plants (Kim et al., 2019). The production of PCs from glutathione is catalyzed by PCS, a protease-like enzyme. In the 30 years since the active PCS from Silene cucubalus was first isolated (Grill et al., 1989), nearly 100 kinds of PCS orPCS-related genes have been identified or predicted in plants, among which the PCS homologs from Pteris vittata (Dong, 2005), Sesbania rostrata (Li et al., 2009), Triticum aestivum (Couselo et al., 2010), tall fescue (Zhao et al., 2014) Oryza sativa (Das et al., 2017), and Ipomoea pes-caprae (Su et al., 2020) have been investigated and found to encode representative PCS proteins. There are single conserved domains at the N- and C-termini in all of these PCS proteins. The conserved N-terminal domain is thought to be a functional center of PCS catalytic activity. The conserved C-terminal domain of unknown function confers metal sensing ability and enzyme stability. In addition, the N-terminal region is more conserved than the C-terminal region (Yazdi et al., 2019; Hayashi et al., 2020). In this study, we isolated the full-length cDNA sequence of BnPCS1 from B. nivea. Sequence alignment showed that BnPCS1 shares high deduced amino acid sequence identities with PCS proteins from other plant species. Two domains, phytochetatin and phytochelatin_C, were found to be present at the N- and C-termini of BnPCS1, respectively. Moreover, the N-terminal domain of BnPCS1 was more conserved than was the C-terminal domain (Figure 1A). The catalytic triad Cys56, His162, and Asp180 consists of three amino acid residues that are essential for the activity of PCS, and these are found in LjPCS1 (Ramos et al., 2007), AtPCS1 (Ha et al., 1999), and PcPCS1 (Hui et al., 2010). BnPCS1 also contains these three specific amino acids, which are Cys58, His164, and Asp182. Cys, as the only amino acid that possesses a sulfhydryl group among the 20 essential amino acids, can bind to heavy metal ions in different forms, which is considered to be important for the activation of PCS. For PCS, being rich in Cys is one of its important features. The C358C359XXXC363XXC366 motif is a heavy metal sensor, which is capable of transferring heavy metal ions to the N-terminal catalytic region when they are detected (Vestergaard et al., 2008; Li et al., 2020). This motif appears as C368C369RETC373MKC376 in LjPCS1 and C369C370QETC374VKC377 in PcPCS1, while this motif is present as C371C372QETC376VKC379 in BnPCS1 (Figure 1A), which is identical to the motif in PcPCS1 and similar to that in LjPCS1. These results reveal that BnPCS1 has similar characteristics to the PCS homologs from the other diverse plant species, and may perform similar functions.
Earlier studies suggested that PCS is a constitutively expressed gene and that there is no regulation of PCS at the transcriptional level in the PC synthesis process. Real-time PCR and Northern blotting results also showed that the transcriptional level of AtPCS1 is not obviously influenced by Cd. Moreover, no significant difference in expression among the different tissues of Arabidopsis was observed (Vatamaniuk et al., 1999), and similar results were observed in tomato (Chen et al., 1997). However, more recent studies have shown that the expression of PCS can be regulated by heavy metal ions. The promoter of AtPCS1 was fused to the GUS reporter gene and transformed into Arabidopsis, and GUS activity in transgenic Arabidopsis seedlings treated with Cd for 5 d increased 2-fold compared to that in untreated plants, although the difference gradually disappeared. This result indicates that the expression of AtPCS1 is positively regulated early in plant development (Lee and Korban, 2002). Under Zn or Cd stress, the relative expression of two phytochelatin synthase genes from Morus alba was induced in root, stem and leaf tissues within 24 h of exposure to the metals, with Cd inducing expression more strongly than did Zn (Fan et al., 2018). The expression level of SoPCS were significantly unregulated in Saccharum officinarum roots under cadmium stress (Yousefi et al., 2018). The MT2 and PCS1 gene expression patterns in Azolla species were significantly induced by the heavy metal treatments (Cu, Zn, Ni, and Cd) (Talebi et al., 2019). In our study, the transcription of BnPCS1 was upregulated significantly in the roots, stems, and leaves of B. nivea seedlings after they were treated with 100 μM Cd2+. This result indicates that the expression of BnPCS1 can be induced by Cd treatment. The increased amount of BnPCS1 mRNA in the leaves was significantly higher than in the roots and stems, which is similar to results for PcPCS1 (Hui et al., 2010) and NnPCS1 (Liu et al., 2012). The possible reasons for this are as follows: (1) B. nivea is a species of herbaceous perennial in the botanical family Urticaceae, and the replacement of the leaves occurs faster than the replacement of roots and stems; (2) the leaves of B. nivea are large with ample, well-developed vacuoles in the mesophyll cells, which can provide enoughbuffer space for Cd2+; (3) many trichomes are distributed on the leaf underside, and trichomes can accumulate heavy metal ions, including Cd2+ (Salt et al., 1995). Abscisic acid (ABA) and salicylic acid (SA) are two important plant hormones that are involved in signal transduction pathways, especially those that play roles in the responses of plants to a multitude of abiotic stresses. The expression of StPCS1 in the roots of potato was strongly induced by exogenous ABA (Stroiński et al., 2010, 2013). Studies on the PCS genes in Arabidopsis showed that exogenous SA has no obvious influence on the transcription of AtPCS1 and AtPCS2 (Cazalé and Clemens, 2001). In this study, the expression of BnPCS1 increased significantly when the B. nivea seedlings were treated with exogenous ABA, while no significant changes in the expression of BnPCS1 were observed when the plants were treated with SA. These results indicate that the expression of BnPCS1 may be independent of the SA signaling pathway, which is similar to the PCS genes from tomato and Arabidopsis.
It is generally accepted that PCS, which catalyzes PC synthesis as a catalytic enzyme, would be localized in the cytoplasm of cells. However, the subcellular localization analysis of BnPCS1 transiently expressed in Arabidopsis protoplasts showed that BnPCS1 was not only distributed in the cytoplasm, but also localized to the nucleus (Figure 2), which is similar to results of previous studies on AtPCS2, a PCS from Arabidopsis that is localized to both the cytoplasm and nucleus (Ding et al., 2013). This result indicates that BnPCS1 may participate in other special physiological processes. For example, some studies have shown that PCs may play important roles in maintaining homeostasis of nutrient ions such as Zn2+ (Tennstedt et al., 2009). Other studies have shown that PC-metal complexes, such as PCs-Cu and PCs-Zn, which function as metal ion donors for metal-dependent enzymes, can activate Cu2+ diamine oxidase (DAO; E.C.1.4.3.6.) and Zn2+ dependent carbonic anhydrase (CA; E.C.4.2.1) (Song et al., 2014).
The functions of PCS enzymes have been investigated through heterologous expression of PCS genes in prokaryotes, yeast, and model plants such as tobacco and Arabidopsis since they were first isolated from various species. However, the results from transgenic plants were various. Arabidopsis plants overexpressing AtPCS1 showed hypersensitivity to Cd stress (Lee et al., 2003b) and enhanced As tolerance (Li et al., 2004), and the accumulation of Cd in the transgenic plants was decreased (Lee et al., 2003a). Heteroexpression of the wheat phytochelatin synthase gene (TaPCS1) in rice enhances cadmium sensitivity (Wang et al., 2012). On the contrary, expression of AtPCS1 in Indian mustard enhanced its tolerance to As and Cd stress (Gasic and Korban, 2007). The accumulation of both As and Cd in tobacco was improved by overexpressing CdPCS1, a phytochelatin synthase gene from Ceratophyllum demersum (Shukla et al., 2012). The OsPCS1 mutants of Oryza sativa exhibited increased sensitivity to As and Cd in hydroponic experiments, showing the importance of OsPCS1-dependent PC synthesis for rice As and Cd tolerance (Uraguchi et al., 2017). Tobacco expressing NtPCS1 from Nelumbo nucifera exhibited an increased tolerance to As and Cd (Lee and Hwang, 2015). Overexpression of three duplicated BnPCS genes from Brassica napus enhanced Cd accumulation and translocationin Arabidopsis thaliana AtPCS1 mutant (Bai et al., 2019). In our study, Arabidopsis seedlings overexpressing BnPCS1 exhibited increased root lengths and fresh weights, but decreased H2O2 and MDA levels. Heavy metal stress can induce ROS such as H2O2 and , which can damage the plasma membrane through lipid peroxidation and other biomolecules such as DNA. MDA is one of the best known secondary products of lipid peroxidation, and it is often used as a marker of cell membrane damage. Compared to WT plants, lower H2O2 and MDA levels in BnPCS1-expressing transgenic Arabidopsis plants suggests that cellular damage was reduced. Cd accumulation in shoots and the translocation factor (TF) of transgenic lines were also found to be significantly higher than those of WT. TF is important for assessing the feasibility of a plant species for phytoremediation purposes. The increased TF indicated that BnPCS1 can enhance the translocation of Cd from roots to shoots in plant. In conclusion, overexpression of BnPCS1 confers enhanced Cd tolerance, accumulation and translocation in transgenic Arabidopsisplants, which could provide gene resources for phytoremediation.
Conclusion
In this study, we isolated a phytochelatin synthase (PCS) gene, BnPCS1, from the bast fiber crop ramie (Boehmeria nivea). Sequence analysis indicated that BnPCS1 encodes a protein of 56.02 kDa that is highly homologous to most of the PCS proteins reported from other plant species. The promoter of BnPCS1 contains several cis-acting elements predicted to be involved in phytohormone signaling and a variety of stress responses. Subcellular localization analysis showed that BnPCS1 localizes to both nucleus and cytoplasm. Real-time PCR analysis showed that BnPCS1 is significantly induced by Cd and ABA. Overexpression lines of BnPCS1 exhibited better root growth and fresh weight, lower level of MDA and H2O2, and higher Cd accumulation and translocation factor compared to the WT under Cd stress. It will be of interest to explore the mechanism how BnPCS1 is regulated by upstream factors, especially through the ABA-dependent pathways and further analysis in the distribution of PC-Cd complexes in plants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SZ designed the study and wrote the manuscript. WS and YJ performed the experiments. All authors discussed and interpreted the results.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- ABA
Abscisic acid
- As
Arsenic
- B. nivea
Boehmeria nivea
- Cd
Cadmium
- cDNA
Complementary DNA
- Cu
Copper
- DW
Dry weight
- Fw
Fresh weight
- GFP
Green fluorescent protein
- GSH
Glutathione
- ORF
Open reading frame
- Pb
Lead
- PCs
Phytochelatins
- PCS
Phytochelatin synthase
- qRT-PCR
Real-time quantitative polymerase chain reaction
- RACE
Rapid amplification of cDNA ends
- SA
Salicylic acid
- TF
Translocation factor
- WT
Wild-type
- Zn
Zinc.
Funding. This study was supported by the National Natural Science Foundation of China (31371704) and the Education Department of Jiangxi province (GJJ190857). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.639189/full#supplementary-material
Nucleotide and deduced amino acid sequence of BnPCS1 from Boehmeria nivea. Nucleotides are numbered on the left. The deduced amino acid residues are shown beneath the corresponding codons. An asterisk indicates the stop codon.
Nucleotide sequence of the BnPCS1 promoter region showing the cis-elements predicted by the PlantCARE database. The BnPCS1 gene promoter sequence is 1,858 bp in length. The “A” of the ATG initiation codon is defined as +1. The CAAT-box, TATA-box, ABRE, ARE, CGTCA-motif, DRE1, G-box, STRE, TGACG-motif, and other important cis-regulatory elements are boxed and labeled.
Kanamycin screening of BnPCS1 transgenic Arabidopsis thaliana seedlings. T0-generation seeds of Arabidopsis thaliana plants transformed with the BnPCS1 gene construct via the floral dip method were germinated on MS medium supplemented with 30 mg⋅L–1 kanamycin for screening. Transgenic lines remain green and grow, while the WT type seedlings stop growing and turn yellow.
Identification of transgenic BnPCS1 Arabidopsis thaliana Col-0 seedlings by amplification of the BsPCS1 gene using the primer pair PCS1-35S-F/PCS1-SP-R. The amplification products were electrophoresed on a 1.5% agarose gel at 5 volt/cm2, 1× TBE buffer for 0.5 h. M: Trans2K Plus II DNA size marker; P: positive plasmid control; CK: no template negative control; WT: Col-0 seedling; L1-L5: BnPCS1 transgenic seedlings. The pBI121-BnPCS1 vector was used to transform Arabidopsis thaliana Col-0 plants via Agrobacterium tumefaciens-mediated transformation. All 35S::BnPCS1-overexpressing transgenic lines (T1 generation) were verified by PCR using the PCS1-35S-F and PCS1-SP-R primers to amplify BcPCS1 from genomic DNA.
Relative expression of the BnPCS1 gene in transgenic Arabidopsis thaliana plants. WT: wild type Arabidopsis Col-0; L1-L5: BnPCS1 transgenic Arabidopsis seedlings; Total RNA was extracted from leaves of transgenic plants overexpressing 35S:: BnPCS1 (T2 generation) for qRT-PCR. BnPCS1 transcript levels were significantly higher in two of the transgenic lines, L1 and L3, which were used in the experiments. Data are presented as the means of three biological replicates with SE shown by vertical bars.
List of oligonucleotide primers used in this study.
References
- Bai J., Wang X., Wang R., Wang J., Le S., Zhao Y. (2019). Overexpression of three duplicated BnPCS genes enhanced cd accumulation and translocation in Arabidopsis thaliana mutant cad1-3. B. Environ. Contam. Tox. 102 146–152. 10.1007/s00128-018-2487-1 [DOI] [PubMed] [Google Scholar]
- Cazalé A. C., Clemens S. (2001). Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS. Lett. 507 215–219. 10.1016/s0014-5793(01)02976-3 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhou J. M., Goldsbrough P. (1997). Characterization of phytochelatin synthase from tomato. Physiol. Plantarum. 101 165–172. 10.1111/j.1399-3054.1997.tb01833.x [DOI] [Google Scholar]
- Choudhury S., Panda P., Sahoo L., Panda S. K. (2013). Reactive oxygen species signaling in plants under abiotic stress. Plant. Signal. Behav. 8:e23681. 10.4161/psb.23681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Couselo J. L., Navarro-Avñó J., Ballester A. (2010). Expression of the phytochelatin synthase TaPCS1 in transgenic aspen, insight into the problems and qualities in phytoremediation of Pb. Int. J. Phytoremediat. 12 358–370. 10.1080/15226510902968134 [DOI] [PubMed] [Google Scholar]
- Das N., Bhattacharya S., Bhattacharyya S., Maiti M. K. (2017). Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol. Biol. 94 167–183. 10.1007/s11103-017-0600-1 [DOI] [PubMed] [Google Scholar]
- DeLong J. M., Prange R. K., Hodges D. M., Forney C. F., Bishop M. C., Quilliam M. (2002). Using a modified ferrous oxidation-xylenol orange (FOX) assay for detection of lipid hydroperoxides in plant tissue. J. Agric. Food Chem. 50 248–254. 10.1021/jf0106695 [DOI] [PubMed] [Google Scholar]
- Ding G., Peng J., Zhang G., Yi H., Fu Y., Gong J. M. (2013). Regulation of the phytochelatin synthase gene AtPCS2 in Arabidopsis thaliana. Scientia Sinica Vitae 43:1112. 10.1360/052013-309 [DOI] [Google Scholar]
- Dong R. (2005). Molecular cloning and characterization of a phytochelatin synthase gene, PvPCS1, from Pteris vittata L. J. Ind. Microbiol. Biot. 32 382–382. 10.1007/s10295-005-0262-x [DOI] [PubMed] [Google Scholar]
- Emamverdian A., Ding Y., Mokhberdoran F., Xie Y. (2015). Heavy metal stress and some mechanisms of plant defense response. Sci. World J. 2015:756120. 10.1155/2015/756120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Guo Q., Liu C., Liu X., Zhang M., Long D., et al. (2018). Two mulberry phytochelatin synthase genes confer zinc/cadmium tolerance and accumulation in transgenic Arabidopsis and tobacco. Gene 645 95–104. 10.1016/j.gene.2017.12.042 [DOI] [PubMed] [Google Scholar]
- Filiz E., Saracoglu I., Ozyigit I., Yalcin B. (2019). Comparative analyses of phytochelatin synthase (PCS) genes in higher plants. Biotechnol. Biotec. Eq. 33 178–194. 10.1080/13102818.2018.1559096 [DOI] [Google Scholar]
- Gasic K., Korban S. S. (2007). Transgenic indian mustard (Brassica juncea) plants expressing an arabidopsis phytochelatin synthase (AtPCS1) exhibit enhanced as and Cd tolerance. Plant Mol. Biol. 64 361–369. 10.1007/s11103-007-9158-7 [DOI] [PubMed] [Google Scholar]
- Grill E., Löffler S., Winnacker E.-L., Zenk M. H. (1989). Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific γ-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc. Natl. Acad. Sci. India B 86:6838. 10.1073/pnas.86.18.6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotto D., Maria L., Valentini J., Paniz C., Schmitt G., Garcia S., et al. (2009). Importance of the lipid peroxidation biomarkers and methodological aspects FOR malondialdehyde quantification. Quim. Nova 32 169–174. 10.1590/S0100-40422009000100032 [DOI] [Google Scholar]
- Ha S. B., Smith A. P., Howden R., Dietrich W. M., Bugg S., O’Connell M. J., et al. (1999). Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11 1153–1164. 10.1105/tpc.11.6.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Tanikawa H., Kuramata M., Abe T., Ishikawa S. (2020). Domain exchange between Oryza sativa phytochelatin synthases reveals a region that determines responsiveness to arsenic and heavy metals. Biochem. Bioph. Res. Co. 523 548–553. 10.1016/j.bbrc.2019.12.093 [DOI] [PubMed] [Google Scholar]
- Hodges D., DeLong J., Forney C., Prange R. (1999). Improving the thiobarbituric acid-reactive-substance assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207 604–611. 10.1007/s004250050524 [DOI] [PubMed] [Google Scholar]
- Hui L., Yu C., Hong-Wei W., Bao-Long S., Jing L., You-Hong C. (2010). Molecular cloning and expression analysis of a phytochelatin synthase gene, PcPCS1, from pyrus calleryana dcne. Acta Horticulturae Sinica 37 880–890. [Google Scholar]
- Kim Y. O., Kang H., Ahn S. J. (2019). Overexpression of phytochelatin synthase AtPCS2 enhances salt tolerance in Arabidopsis thaliana. J. Plant Physiol. 240:153011. 10.1016/j.jplph.2019.153011 [DOI] [PubMed] [Google Scholar]
- Lee B. D., Hwang S. (2015). Tobacco phytochelatin synthase (NtPCS1) plays important roles in cadmium and arsenic tolerance and in early plant development in tobacco. Plant Biotechnol. Rep. 9 107–114. 10.1007/s11816-015-0348-5 [DOI] [Google Scholar]
- Lee J., Bae H., Jeong J., Lee J. Y., Yang Y. Y., Hwang I., et al. (2003a). Functional expression of a bacterial heavy metal transporter in Arabidopsis enhances resistance to and decreases uptake of heavy metals. Plant Physiol. 133 589–596. 10.1104/pp.103.021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Korban S. S. (2002). Transcriptional regulation of Arabidopsis thaliana phytochelatin synthase (AtPCS1) by cadmium during early stages of plant development. Planta 215 689–693. 10.1007/s00425-002-0821-6 [DOI] [PubMed] [Google Scholar]
- Lee S., Moon J. S., Ko T. S., Petros D., Goldsbrough P. B., Korban S. S. (2003b). Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 131 656–663. 10.1104/pp.014118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. M., Yu B. Y., Chen F. H., Gan H. Y., Yuan J. G., Qiu R., et al. (2009). Characterization of the Sesbania rostrata phytochelatin synthase gene: alternative splicing and function of four isoforms. Int. J. Mol. Sci. 10 3269–3282. 10.3390/ijms10083269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Barbaro E., Bellini E., Saba A., Sanità di Toppi L., Varotto C. (2020). Ancestral function of the phytochelatin synthase C-terminal domain in inhibition of heavy metal-mediated enzyme overactivation. J. Exp. Bot. 71 6655–6669. 10.1093/jxb/eraa386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dhankher O. P., Carreira L., Lee D., Chen A., Schroeder J. I., et al. (2004). Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant Cell Physiol. 45 1787–1797. 10.1093/pcp/pch202 [DOI] [PubMed] [Google Scholar]
- Liu Z., Gu C., Chen F., Yang D., Wu K., Chen S., et al. (2012). Heterologous expression of a Nelumbo nucifera phytochelatin synthase gene enhances cadmium tolerance in Arabidopsis thaliana. Appl. Biochem. Biotechnol. 166 722–734. 10.1007/s12010-011-9461-2 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum 15 437–496. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Pomponi M., Censi V., Di Girolamo V., De Paolis A., di Toppi L. S., Aromolo R., et al. (2006). Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd2+ tolerance and accumulation but not translocation to the shoot. Planta 223 180–190. 10.1007/s00425-005-0073-3 [DOI] [PubMed] [Google Scholar]
- Ramos J., Clemente M. R., Naya L., Loscos J., Pérez-Rontomé C., Sato S., et al. (2007). Phytochelatin synthases of the model legume Lotus japonicus. a small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiol. 143 1110–1118. 10.1104/pp.106.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigouin C., Nylin E., Cogswell A. A., Schaumlöffel D., Dobritzsch D., Williams D. L. (2013). Towards an understanding of the function of the phytochelatin synthase of Schistosoma mansoni. PLoS. Negl. Trop. Dis. 7:e2037. 10.1371/journal.pntd.0002037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt D. E., Prince R. C., Pickering I. J., Raskin I. (1995). Mechanisms of cadmium mobility and accumulation in indian mustard. Plant Physiol. 109 1427–1433. 10.1104/pp.109.4.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W., Jie Y., Xing H., Lu Y., Kang W., Wang D. (2011). Heavy metal concentrations and bioaccumulation of ramie (Boehmeria nivea) growing on 3 mining areas in shimen, lengshuijiang and Liuyang of Hunan province. Acta Ecologica. Sinica 31 874–881. [Google Scholar]
- She W., Zhu S., Jie Y., Xing H., Cui G. (2015). Expression profiling of cadmium response genes in ramie (Boehmeria nivea L.) root. Bull. Environ. Contam. Toxicol. 94 453–459. 10.1007/s00128-015-1502-z [DOI] [PubMed] [Google Scholar]
- Shukla D., Kesari R., Mishra S., Dwivedi S., Tripathi R. D., Nath P., et al. (2012). Expression of phytochelatin synthase from aquatic macrophyte Ceratophyllum demersum L. enhances cadmium and arsenic accumulation in tobacco. Plant Cell Rep. 31 1687–1699. 10.1007/s00299-012-1283-3 [DOI] [PubMed] [Google Scholar]
- Song W. Y., Mendoza-Cózatl D. G., Lee Y., Schroeder J. I., Ahn S. N., Lee H. S., et al. (2014). Phytochelatin-metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Environ. 37 1192–1201. 10.1111/pce.12227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroiński A., Chadzinikolau T., Giżewska K., Zielezińska M. (2010). ABA or cadmium induced phytochelatin synthesis in potato tubers. Biol. Plantarum. 54 117–120. 10.1007/s10535-010-0017-z [DOI] [Google Scholar]
- Stroiński A., Giżewska K., Zielezińska M. (2013). Abscisic acid is required in transduction of cadmium signal to potato roots. Biol. Plantarum. 57 121–127. 10.1007/s10535-012-0135-x [DOI] [Google Scholar]
- Su H., Zou T., Lin R., Zheng J., Jian S., Zhang M. (2020). Characterization of a phytochelatin synthase gene from ipomoea pes-caprae involved in cadmium tolerance and accumulation in yeast and plants. Plant Physiol. Biochem. 155 743–755. 10.1016/j.plaphy.2020.08.012 [DOI] [PubMed] [Google Scholar]
- Talebi M., Tabatabaei B. E. S., Akbarzadeh H. (2019). Hyperaccumulation of Cu, Zn, Ni, and Cd in Azolla species inducing expression of methallothionein and phytochelatin synthase genes. Chemosphere 230 488–497. 10.1016/j.chemosphere.2019.05.098 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tennstedt P., Peisker D., Böttcher C., Trampczynska A., Clemens S. (2009). Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiol. 149 938–948. 10.1104/pp.108.127472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N. (1980). Limiting factors in photosynthesis. 1. Use of iron stress to control photochemical capacity in vivo. Plant Physiol. 65 114–120. 10.1104/pp.65.1.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S., Tanaka N., Hofmann C., Abiko K., Ohkama-Ohtsu N., Weber M., et al. (2017). Phytochelatin synthase has contrasting effects on cadmium and arsenic accumulation in rice grains. Plant Cell Physiol. 58 1730–1742. 10.1093/pcp/pcx114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk O. K., Mari S., Lang A., Chalasani S., Demkiv L. O., Rea P. A. (2004). Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with gamma-glutamylcysteine during catalysis: stoichiometric and site-directed mutagenic analysis of Arabidopsis thaliana PCS1-catalyzed phytochelatin synthesis. J. Biol. Chem. 279 22449–22460. 10.1074/jbc.M313142200 [DOI] [PubMed] [Google Scholar]
- Vatamaniuk O. K., Mari S., Lu Y.-P., Rea P. A. (1999). AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. India B 96:7110. 10.1073/pnas.96.12.7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard M., Matsumoto S., Nishikori S., Shiraki K., Hirata K., Takagi M. (2008). Chelation of cadmium ions by phytochelatin synthase: role of the cysteine-rich C-terminal. Anal. Sci. 24 277–281. 10.2116/analsci.24.277 [DOI] [PubMed] [Google Scholar]
- Wang F., Wang Z., Zhu C. (2012). Heteroexpression of the wheat phytochelatin synthase gene (TaPCS1) in rice enhances cadmium sensitivity. Acta Biochim. Biophys. Sin. 44 886–893. 10.1093/abbs/gms073 [DOI] [PubMed] [Google Scholar]
- Yazdi M., Kolahi M., Mohajel Kazemi E., Goldson Barnaby A. (2019). Study of the contamination rate and change in growth features of lettuce (Lactuca sativa Linn.) in response to cadmium and a survey of its phytochelatin synthase gene. Ecotoxicol. Environ. Saf. 180 295–308. 10.1016/j.ecoenv.2019.04.071 [DOI] [PubMed] [Google Scholar]
- Yoo S., Cho Y., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yousefi Z., Kolahi M., Majd A., Jonoubi P. (2018). Effect of cadmium on morphometric traits, antioxidant enzyme activity and phytochelatin synthase gene expression (SoPCS) of saccharum officinarum var. cp48-103 in vitro. Ecotoxicol. Environ. Saf. 157 472–481. 10.1016/j.ecoenv.2018.03.076 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1 641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhao C., Xu J., Li Q., Li S., Wang P., Xiang F. (2014). Cloning and characterization of a Phragmites australis phytochelatin synthase (PaPCS) and achieving Cd tolerance in tall fescue. PLoS One 9:e103771. 10.1371/journal.pone.0103771 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide and deduced amino acid sequence of BnPCS1 from Boehmeria nivea. Nucleotides are numbered on the left. The deduced amino acid residues are shown beneath the corresponding codons. An asterisk indicates the stop codon.
Nucleotide sequence of the BnPCS1 promoter region showing the cis-elements predicted by the PlantCARE database. The BnPCS1 gene promoter sequence is 1,858 bp in length. The “A” of the ATG initiation codon is defined as +1. The CAAT-box, TATA-box, ABRE, ARE, CGTCA-motif, DRE1, G-box, STRE, TGACG-motif, and other important cis-regulatory elements are boxed and labeled.
Kanamycin screening of BnPCS1 transgenic Arabidopsis thaliana seedlings. T0-generation seeds of Arabidopsis thaliana plants transformed with the BnPCS1 gene construct via the floral dip method were germinated on MS medium supplemented with 30 mg⋅L–1 kanamycin for screening. Transgenic lines remain green and grow, while the WT type seedlings stop growing and turn yellow.
Identification of transgenic BnPCS1 Arabidopsis thaliana Col-0 seedlings by amplification of the BsPCS1 gene using the primer pair PCS1-35S-F/PCS1-SP-R. The amplification products were electrophoresed on a 1.5% agarose gel at 5 volt/cm2, 1× TBE buffer for 0.5 h. M: Trans2K Plus II DNA size marker; P: positive plasmid control; CK: no template negative control; WT: Col-0 seedling; L1-L5: BnPCS1 transgenic seedlings. The pBI121-BnPCS1 vector was used to transform Arabidopsis thaliana Col-0 plants via Agrobacterium tumefaciens-mediated transformation. All 35S::BnPCS1-overexpressing transgenic lines (T1 generation) were verified by PCR using the PCS1-35S-F and PCS1-SP-R primers to amplify BcPCS1 from genomic DNA.
Relative expression of the BnPCS1 gene in transgenic Arabidopsis thaliana plants. WT: wild type Arabidopsis Col-0; L1-L5: BnPCS1 transgenic Arabidopsis seedlings; Total RNA was extracted from leaves of transgenic plants overexpressing 35S:: BnPCS1 (T2 generation) for qRT-PCR. BnPCS1 transcript levels were significantly higher in two of the transgenic lines, L1 and L3, which were used in the experiments. Data are presented as the means of three biological replicates with SE shown by vertical bars.
List of oligonucleotide primers used in this study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.