Fig 1.
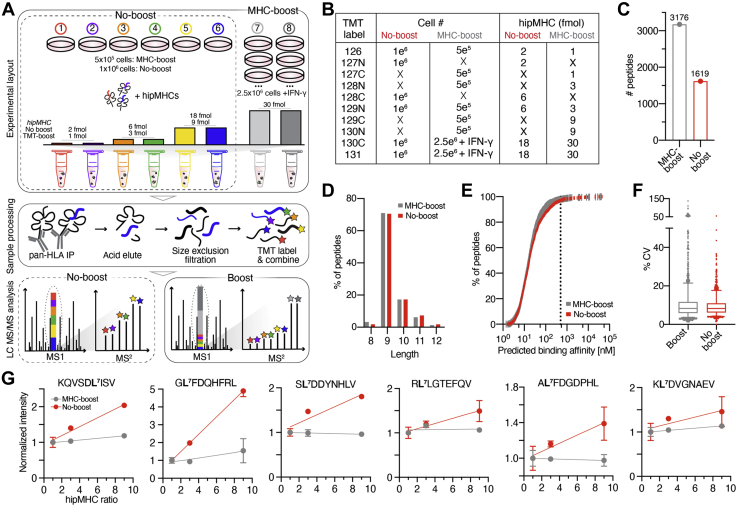
Estimating ratio compression using hipMHCs in immunopeptidomic analyses.A, experimental setup of hipMHC quantitative immunopeptidomic analyses ± protein carrier. B, experimental setup of isobaric labels, cell number, and concentration of hipMHC added for each sample. C, number of unique pMHCs identified in a single analysis. D, length distribution of pMHCs. E, predicted binding affinity of 9-mers. 97.9% and 97.0% of 9-mers in the pMHC-boost and no-boost analyses, respectively, were predicted to have a binding affinity ≤500 nM (dotted line). F, coefficients of variation of pMHC-boost and no-boost analyses. Boxes outline the interquartile range, and whiskers the 5 and 95th percentiles. pMHC-boost median CV = 8.23%, no-boost = 8.30%. 95% PSMs have CV <17.7% (no-boost) and 21.5% (pMHC-boost), Violin plots of reporter ion intensities for pMHC-boost (left) and no-boost (right) analyses. Median: black dashed line, quartiles: colored dotted line. G, reporter ion intensities of hipMHC peptides normalized to the mean signal of the 1 fmol (pMHC-boost) or 2 fmol (no-boost) samples (y-axis), where the hipMHC ratio represents the amount of hipMHC added over the lowest concentration (x-axis). Solid line = linear fit, error bars show ± standard deviation. Source data can be found in supplemental Table S3.