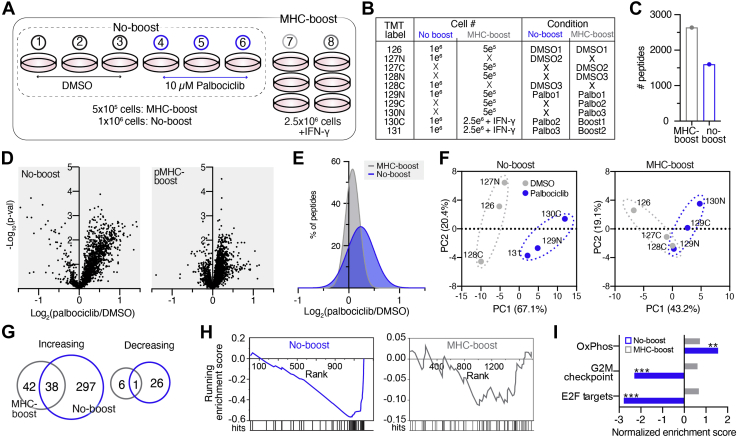
Fig 2.
Palbociclib-induced pMHC repertoire alterations are masked in the presence of a protein carrier.A, experimental setup of pMHC analyses ± protein carrier with 72 h DMSO or 10 μM palbociclib treatment. B, experimental setup of isobaric labels, cell number, and treatment conditions. C, number of unique peptides identified in pMHC-boost (2637) and no-boost (1602) analyses. D, volcano plot displaying the log2(palbociclib/DMSO) of pMHCs (x-axis), where the fold change is calculated from the mean intensity of n = 3 biological replicates per condition, versus significance (y-axis, mean adjusted p-value, unpaired two-sided t test). E, histogram distribution of unique pMHC fold change in expression. F, samples plotted by principal component 1 (PC1) and PC2 score for no-boost (left) and pMHC-boost (right) analysis, colored by treatment condition. Percentages are % variance explained by the plotted PC. G, Venn diagram of peptides significantly increasing (upper) and decreasing (lower) with palbociclib treatment in the no-boost (blue) and pMHC-boost (gray) analyses. H, pMHC enrichment plots for E2F targets for the no-boost (gray, p = 0.13, q = 0.88) and pMHC-boost (blue, p < 0.001, q < 0.001) analyses. Hits mark pMHCs of source proteins mapping to E2F targets. I, normalized enrichment scores from enrichment analyses of pMHC-boost (gray) and no-boost (blue) datasets. Positive/negative scores represent directionality of pathway enrichment. Significant enrichment is noted by ∗∗p < 0.01, ∗∗∗p < 0.001, with FDR-q values <0.25.