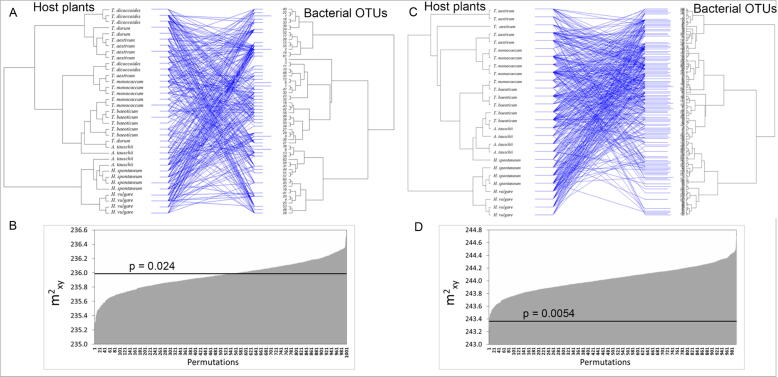
Fig. 7.
Co-evolution analysis. (A) Tanglegram between the cereal phylogeny, based on RAPD analysis, and the bacterial phylogeny (at OTU level), based on the metabarcoding analysis. Blue lines indicate plant–bacterial associations that were more significant than expected by chance, according to the ParaFitGlobal statistic. (B) Visualization of the global goodness-of-fit statistics over 1000 permutation (P = 0.024); the black line indicates the observed global sum of squared residuals (m2XY) value, while the grey columns indicate the m2XY value of each of the 1000 randomizations, sorted by ascending values. (C) and (D) Same than (A) and (B), calculated after removal of T. durum and T. dicoccoides samples.