Fig. 3 |. Neandertal and Denisovan variant depletion varies between enhancers active in different tissues.
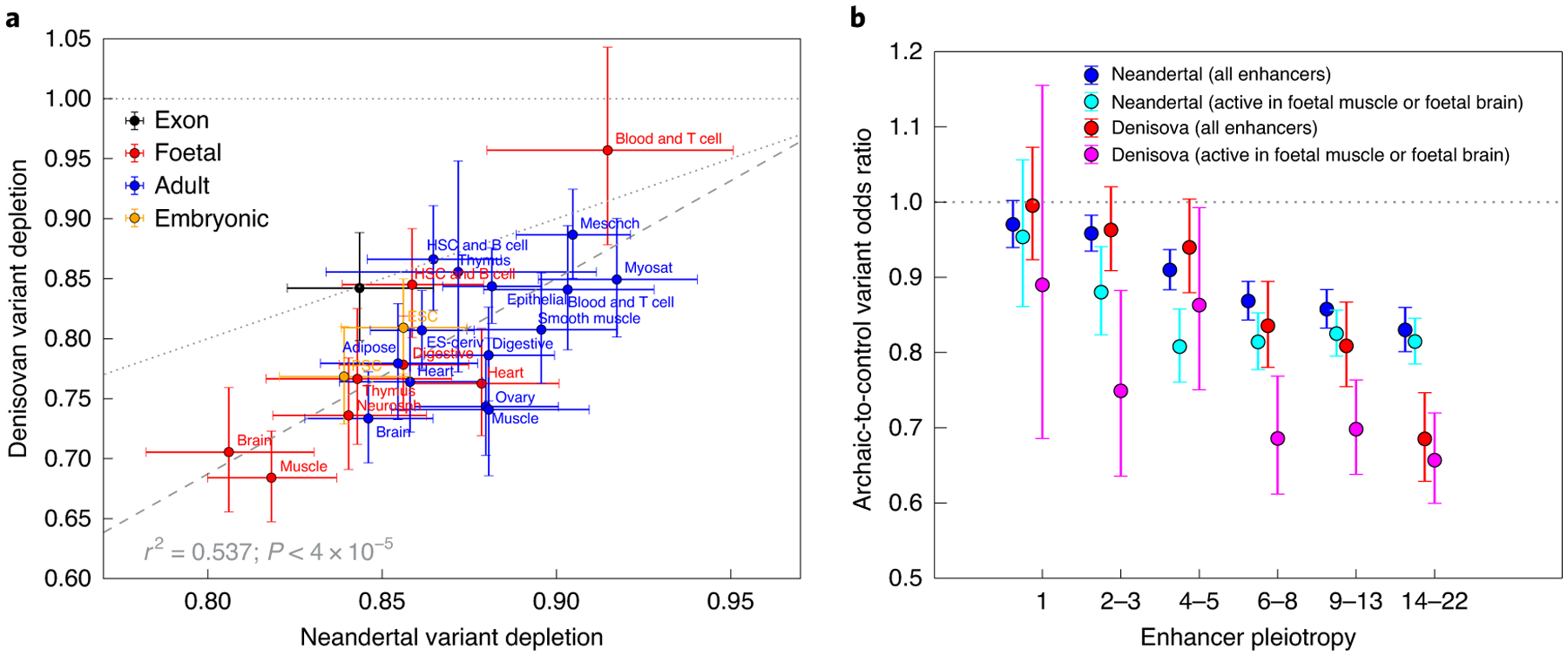
a, The set of enhancers active in each Roadmap cell line is significantly depleted of both Neandertal and Denisovan variation, with the exception of blood and T cells, whose Denisovan depletion confidence interval overlaps an odds ratio of 1.0 (horizontal dotted line). Data points that lie below the diagonal dotted line correspond to tissues whose enhancers are more depleted of Denisovan SNPs compared with Neandertal SNPs. The slope of the dashed regression line is significantly positive (P < 4 × 10−5), implying that Neandertal variant depletion and Denisovan variant depletion are correlated across tissues (r2 = 0.537). b, even after restricting to enhancers active in foetal muscle or foetal brain (the two tissue types most depleted of introgressed variation), pleiotropy remains negatively correlated with archaic SNP depletion. The difference between these two tissues and other tissues is driven mainly by enhancers of intermediate pleiotropy. All error bars span 95% binomial confidence intervals. eSC, embryonic stem cell; HSC, haematopoietic stem cell; mesench, mesenchymal; myosat, myosatellite cell; iPSC, induced pluripotent stem cell; eS-deriv, cell line derived from embryonic stem cells; neurosph, neurosphere cell.
