Extended Data Fig. 2 |. Human versus archaic reference sequence divergence as a function of enhancer pleiotropy and tissue activity.
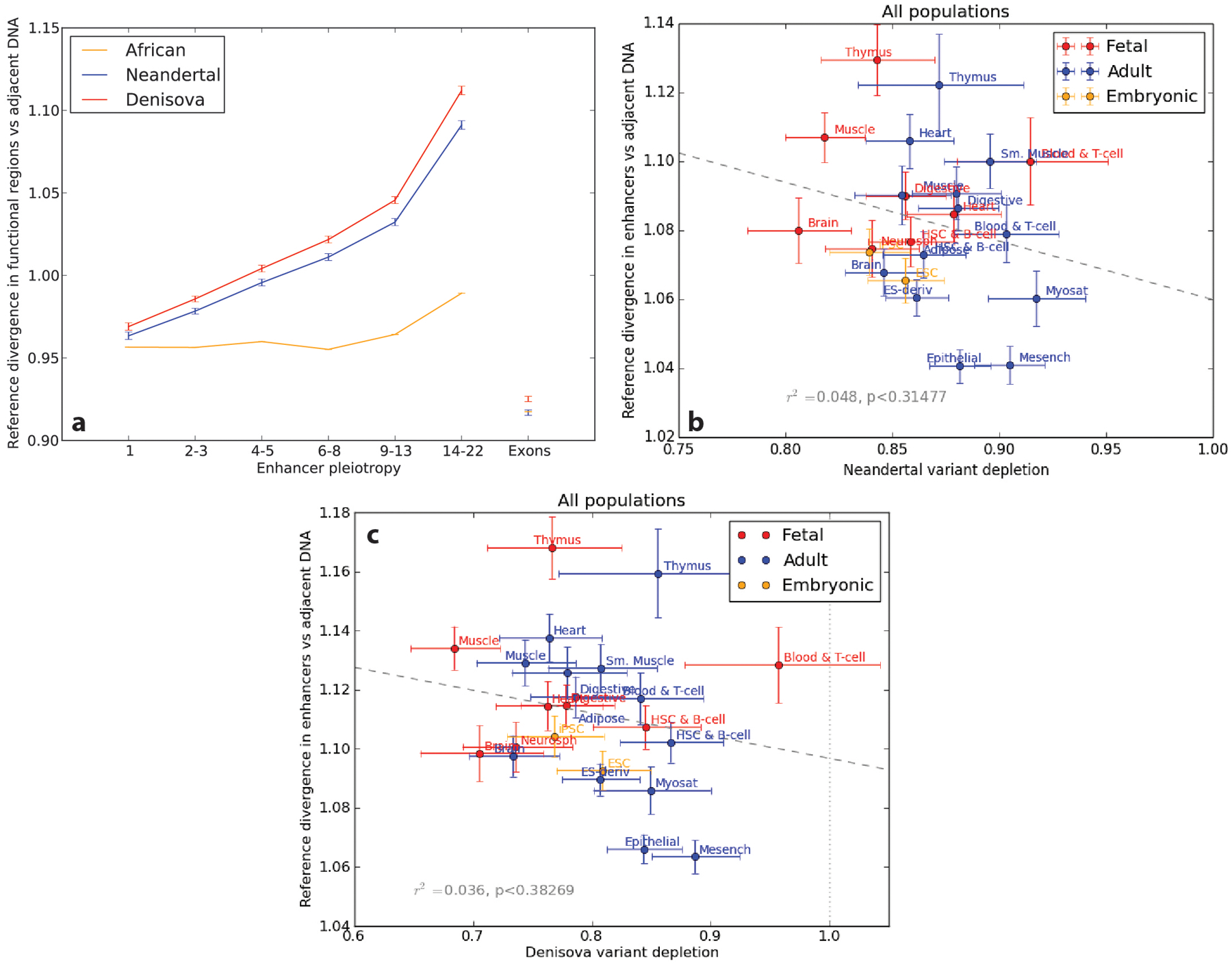
a, Within enhancers and exons, we measured divergence of the human reference from the Altai Neandertal, the Altai Denisovan, and the YRI African genomes, then normalized each by the divergence of the same genomes within adjacent control regions. exon divergence ratios <1 indicate that purifying selection has slowed down their sequence evolution compared to less constrained adjacent regions. In contrast, divergence is accelerated in enhancers relative to control regions. This acceleration is positively correlated with pleiotropy and is stronger for archaic vs. human comparisons than for the African vs. human reference genome comparison. In the absence of selection against archaic enhancer variation, this divergence pattern should cause archaic SNPs to be enriched within high-pleiotropy enhancers, not depleted as we in fact observe. b, We see no correlation across tissue-specific enhancer sets between Neandertal divergence from the human reference and archaic allele depletion from enhancers. This suggests that differences between tissues in the depletion of introgressed archaic variants are not driven by differences in divergence between reference genomes. c, We see no correlation across tissue-specific enhancer sets between Denisovan divergence from the human reference and archaic allele depletion from enhancers. All error bars in panels a–c are 95% confidence intervals derived from the binomial approximation to the Bernoulli distribution.
