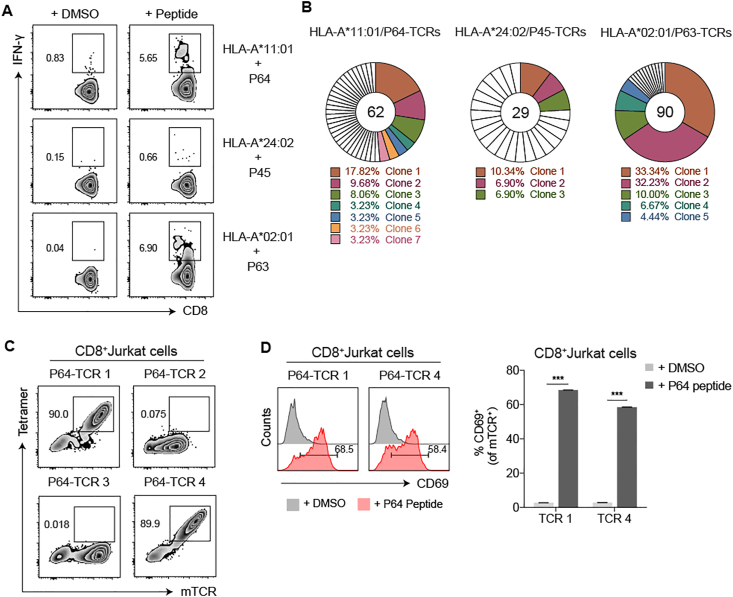
Figure 2.
The isolation and validation of the SARS-CoV-2 specific TCRs of CD8+ T cells. (A) After stimulated by peptide mixtures and expanded for 10 days, single IFN-γ secreting T cell from three COVID-19 convalescent PBMC samples (C40: HLA-A∗11:01+, C33: HLA-A24+ and C27: HLA-A2+) were individually stimulated by P64, P45 and P63, from three corresponding HLA subtypes, and isolated by flow cytometric sorter. (B) The TCRs of sorted cells were amplified by RT-PCR and the TCR repertoire was analyzed with the IMGT/V-Questtool (http://www.imgt.org/) to obtain dominant TCR clones. Unique clones were marked in white. Different colors represented relative dominant clones (copy≧2). (C,D) Specificity verification for the top 4 P64-TCR clones in CD8+ Jurkat cells. P64-HLA-A∗11:01 tetramer (Tetramer) and anti-mouse TCR (mTCR) antibody were applied to evaluate the specific binding of P64-TCRs (C). CD69 was used to determine the specific activation of TCR-transduced CD8+ Jurkat cells, after co-cultured with HLA-A∗11:01 expressingCOS-7 cells pulsed with P64 (D). Grey represented DMSO-stimulated samples and pink represented P64-stimulated samples. Data were presented as mean ± SD, n = 3, ∗∗∗P < 0.001. Representative data of two independent experiments were shown.