Huang et al. reveal that the microcephaly-related protein WDR62 possesses an intrinsic affinity for curved microtubules and regulates spindle dynamics by functioning as an adaptor protein between its recruiting factor TPX2/Aurora A and the effector katanin.
Abstract
WDR62 is a microcephaly-related, microtubule (MT)-associated protein (MAP) that localizes to the spindle pole and regulates spindle organization, but the underlying mechanisms remain elusive. Here, we show that WDR62 regulates spindle dynamics by recruiting katanin to the spindle pole and further reveal a TPX2–Aurora A–WDR62–katanin axis in cells. By combining cellular and in vitro experiments, we demonstrate that WDR62 shows preference for curved segments of dynamic GDP-MTs, as well as GMPCPP- and paclitaxel-stabilized MTs, suggesting that it recognizes extended MT lattice. Consistent with this property, WDR62 alone is inefficient in recruiting katanin to GDP-MTs, while WDR62 complexed with TPX2/Aurora A can potently promote katanin-mediated severing of GDP-MTs in vitro. In addition, the MT-binding affinity of WDR62 is autoinhibited through JNK phosphorylation-induced intramolecular interaction. We propose that WDR62 is an atypical MAP and functions as an adaptor protein between its recruiting factor TPX2/Aurora A and the effector katanin to orchestrate the regulation of spindle dynamics.
Introduction
The mitotic spindle is composed of bipolar arrays of microtubules (MTs) with the minus ends embedded at the spindle pole and the plus ends projecting toward the chromosome. MTs are highly dynamic structures that undergo continual switching between growth and shrinkage. The dynamic changes in the spindle MTs, which are orchestrated by a variety of microtubule-associated proteins (MAPs), play an essential role in ensuring faithful chromosome segregation during mitosis (Petry, 2016; Kapoor, 2017). However, despite a wealth of knowledge on the regulation of the plus-end dynamics of spindle MTs, little is known about how the minus-end dynamics are regulated by the spindle pole–associated MAPs. Interestingly, a significant number of spindle pole–associated proteins, including WDR62 (WD repeat-containing protein 62), have been linked to MCPH (autosomal-recessive primary microcephaly), a neurodevelopmental disorder characterized by a reduction in brain size, highlighting the importance of mitotic spindle regulation in MCPH etiology (Bilgüvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010).
Mutations in WDR62 are the second-most common genetic cause of MCPH (Bilgüvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010). Studies in different model systems have revealed that WDR62 participates in many biological events, including mitotic progression, meiotic initiation, neuronal migration, and proliferation, which may depend on its diverse functions in spindle orientation, spindle assembly, asymmetric centrosome inheritance and maintenance, and centriole duplication, as well as cilium disassembly (Bogoyevitch et al., 2012; Novorol et al., 2013; Chen et al., 2014; Xu et al., 2014; Kodani et al., 2015; Lim et al., 2015; Jayaraman et al., 2016; Ramdas Nair et al., 2016; Sgourdou et al., 2017; Xu et al., 2018; Zhou et al., 2018; Hu and Jasper, 2019; Qin et al., 2019; Zhang et al., 2019). Among these, the spindle-related function has been implicated to rely on its MT-binding activity (Lim et al., 2015). However, the biochemical nature of WDR62’s MT-binding activity is yet obscure, and how this activity contributes to its cellular function remains elusive. The spindle pole localization of WDR62 depends on Aurora A, to which WDR62 directly binds, and TPX2, the upstream activator of Aurora A, demonstrating that WDR62 acts downstream of TPX2/Aurora A (Lim et al., 2015; Lim et al., 2016). However, the exact roles of this pathway in spindle organization and the underlying mechanisms remain unclear.
In this study, we identify katanin, a MT-severing enzyme (Sharp and Ross, 2012; McNally and Roll-Mecak, 2018), as a new binding partner of WDR62. WDR62 recruits katanin to the spindle pole and regulates spindle minus-end depolymerization. WDR62 preferentially binds to curved segments of dynamic MTs, as well as GMPCPP- and paclitaxel-stabilized MTs, suggesting that WDR62 recognizes extended MT lattice. Consistently, WDR62 can recruit katanin to sever curved MT lattices in cells and GMPCPP-stabilized MTs in vitro. Using the auxin-inducible degron (AID) system, we establish in cells a TPX2–Aurora A–WDR62–katanin axis. Reconstitution experiments demonstrated that the TPX2–Aurora A–WDR62–katanin module can potently disassemble GDP-MTs in vitro. Moreover, we reveal that the MT-binding affinity of WDR62 is repressed by an intramolecular interaction mediated by JNK phosphorylation. In summary, we propose that WDR62 recognizes extended MT lattice and regulates spindle dynamics through a TPX2–Aurora A–WDR62–katanin axis and that perturbation of these activities might lead to microcephaly.
Results
WDR62 and katanin form a complex at the spindle pole
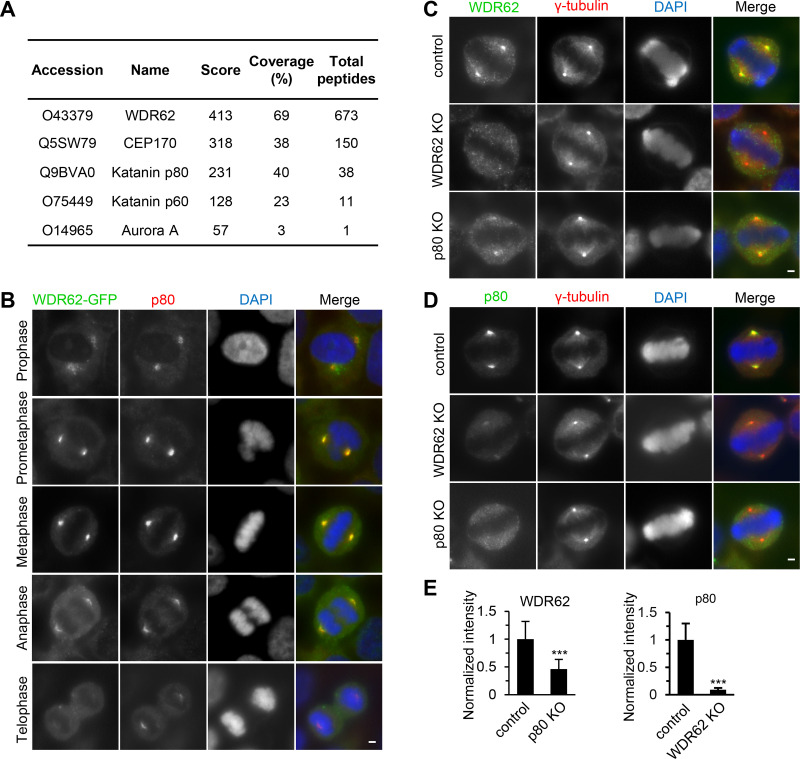
To gain insight into the function of WDR62, we searched for its binding partners. Extracts of HEK293T cells expressing WDR62 with a GFP and biotinylation tag (GFP-Bio) together with the biotin ligase BirA were used for streptavidin pull-down assays, and the resulting proteins were analyzed by mass spectrometry. In addition to the already known binding partners of WDR62, such as CEP170 and Aurora A (Chen et al., 2014; Qin et al., 2019; Zhang et al., 2019), we identified two subunits of katanin, p60 and p80, among the most prominent hits (Fig. 1 A).
Figure 1.
WDR62 interacts with katanin at spindle poles. (A) Mass spectrometry results of streptavidin pull down from HEK293T cells transiently transfected with WDR62-GFP-Bio and synchronized in mitosis by nocodazole treatment. (B) Immunofluorescence staining of p80 and DNA (DAPI) in WDR62-GFP-Strep knock-in HeLa cells during mitosis. (C–E) Immunofluorescence staining and quantification of WDR62 and p80 intensities at spindle poles in control or indicated knockout (KO) HeLa cells. For WDR62 intensity, control, n = 31 spindle poles; p80 knockout, n = 44. For p80 intensity, control, n = 34; WDR62 knockout, n = 47. Scale bars, 2 µm. Data represent mean ± SD. ***, P < 0.001; two-tailed t test.
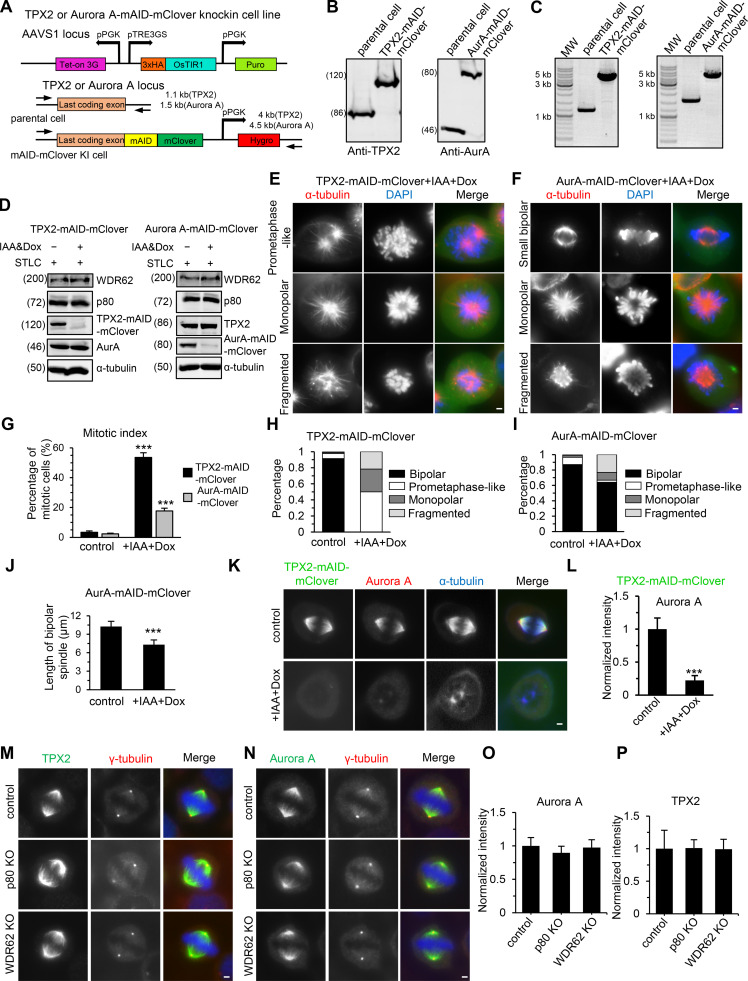
Both WDR62 and katanin have been previously reported to localize to the spindle pole (Nicholas et al., 2010; Bogoyevitch et al., 2012). To compare their subcellular localization directly, we generated a CRISPR knock-in HeLa cell line in which the endogenous WDR62 protein was C-terminally tagged with GFP (Fig. S1, A–C). Immunofluorescent staining in this cell line with an anti-p80 antibody showed that WDR62 and katanin colocalize at spindle poles during mitosis, with the strongest overlap from prometaphase to metaphase (Fig. 1 B). Knockout of WDR62 in HeLa cells dramatically reduced the spindle pole localization of p80 by ∼90%, while knockout of p80 reduced the spindle pole localization of WDR62 by ∼50% (Fig. 1, C–E; and Fig. S1, D and E), demonstrating that WDR62 and katanin depend on each other for their localization and that the dependence of katanin localization on WDR62 is much stronger than vice versa.
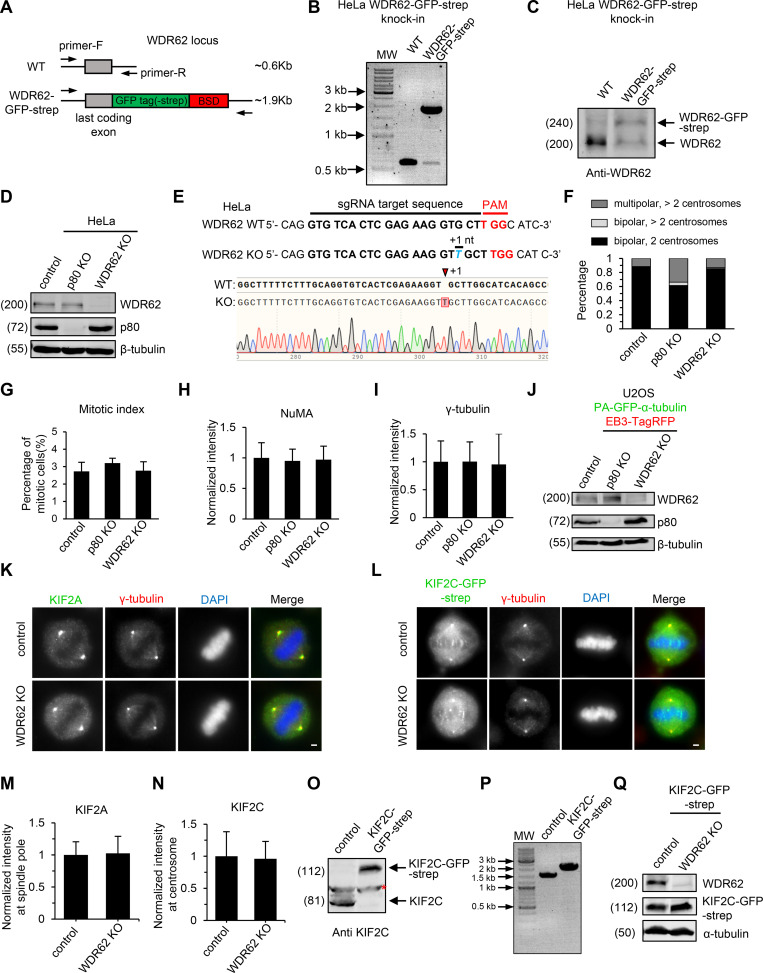
Figure S1.
Characterization of WDR62 knock-in and knockout cell lines.(A–C) Characterization of WD62 knock-in HeLa cell line. (A) Schematic illustration of primer sets and the expected PCR products. PCR genotyping (B) and Western blotting analysis (C) showing that the WDR62-GFP-Strep knock-in HeLa cell line used in this study is heterozygous. BSD, blasticidin S deaminase. (D) Western blotting with the indicated antibodies in control, katanin p80 knockout (KO), and WDR62 knockout HeLa cells. (E) Sequencing results of WDR62 knockout HeLa cell line used in this study. 1 nucleotide (nt) insertion will result in p.V65VfsX13. sgRNA, single-guide RNA. (F) Frequency of multipolarity in control or the indicated knockout HeLa cells in prometaphase or metaphase. Control, n = 224 cells; p80 knockout, n = 189; WDR62 knockout, n = 211. (G–I) Quantification of mitotic index (G), NuMA total intensity (H), and γ-tubulin total intensity (I) in control, p80 knockout, and WDR62 knockout HeLa cells. For mitotic index, n = 3 experiments, control, 1,363 cells; p80 knockout, 1,708 cells; WDR62 knockout, 1,385 cells. For NuMA intensity, n = 70 spindle poles in all conditions. For γ-tubulin intensity, n = 74 centrosomes in all conditions. (J) Western blotting with the indicated antibodies in control, katanin p80 knockout, and WDR62 knockout U2OS stable cell lines expressing PA-GFP–α-tubulin. Both knockouts are from a mixed population of cells after transient transfection with PX459 bearing single guide RNA) followed by drug selection, rather than from single cell cloning. (K and M) Immunofluorescence staining for KIF2A, γ-tubulin, and DAPI (K) and quantification of KIF2A intensity at spindle poles (M) in control and WDR62 knockout HeLa cells. n = 50 spindles poles for both conditions. (L and N) Immunofluorescence staining for γ-tubulin and DAPI (L) and quantification of KIF2C intensity at centrosomes (N) in control and WDR62 knockout KIF2C-GFP-Strep knock-in HeLa cells. The WDR62 knockout is from a mixed population of cells after transient transfection with PX459 bearing single guide RNA followed by drug selection, rather than from single cell cloning. n = 50 spindles poles for both conditions. (O and P) Western blotting (O) and PCR genotyping (P) showing that the KIF2C-GFP-Strep knock-in HeLa cell line used in L is homozygous. Red asterisk denotes a nonspecific band detected by the KIF2C antibody. (Q) Western blotting with the indicated antibodies in control and WDR62 knockout KIF2C-GFP-Strep knock-in HeLa cells. The WDR62 knockout is from a mixed population of cells after transient transfection with PX459 bearing single guide RNA followed by drug selection, rather than from single cell cloning. Scale bars, 2 µm. Data represent mean ± SD.
WDR62-katanin regulates spindle organization and dynamics
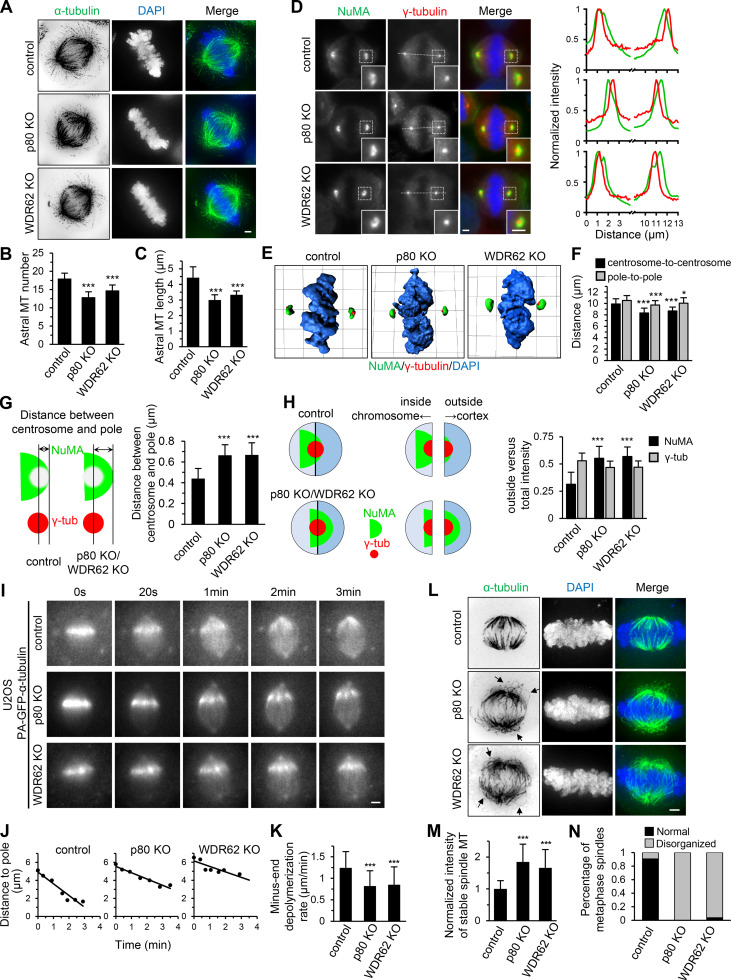
We next investigated the functional significance of the interaction between WDR62 and katanin. Knockout of WDR62 and p80 in HeLa cells decreased both the number and length of astral MTs, as previously described (Fig. 2, A–C; Jiang et al., 2017; Miyamoto et al., 2017). However, unlike p80 knockout, knockout of WDR62 did not cause an increase in extra centrosomes and multipolar spindles (Fig. S1 F), suggesting that katanin’s function in limiting centrosome number and multipolarity is not through WDR62 but other, yet unknown proteins. In addition, the mitotic index did not significantly increase in both knockout cells (Fig. S1 G). We further investigated the centrosome–spindle pole connection by costaining γ-tubulin (centrosome marker) and NuMA (spindle pole marker). In both WDR62 and p80 knockout HeLa cells, the centrosome exhibited an inward displacement relative to the spindle pole, making the centrosome appear “embedded” within the spindle pole, as determined by the immunofluorescence images and the shifted line scan intensity profiles as well as 3D reconstructions (Fig. 2, D and E). Quantitative analyzes revealed that compared with control metaphase spindles, the pole-to-pole distance was slightly decreased by 5% and 8% while the centrosome-to-centrosome distance was decreased by 12% and 16% in WDR62 knockout and p80 knockout cells, respectively (Fig. 2 F); accordingly, the distance between centrosome and spindle pole (Fig. 2 G, denoted by double-headed arrow) increased ∼1.5-fold in both knockout cells. Consistently, although the total intensity of γ-tubulin and NuMA remained largely unchanged (Fig. S1, H and I), the ratio of “outside” versus total NuMA intensity increased approximately twofold in both knockout cells (Fig. 2 H; Materials and methods for details).
Figure 2.
WDR62-katanin regulates spindle organization and dynamics.(A) Immunofluorescence staining for α-tubulin and DAPI in control and indicated knockout (KO) HeLa cells. Maximum intensity projections of Z series with 30 stacks at 0.11-µm steps are shown. (B and C) Quantification of astral MT number and length as shown in A. n = 28, 24, and 20 spindle poles for control, p80 knockout, and WDR62 knockout, respectively. (D) Left: Immunofluorescence staining for NuMA, γ-tubulin, and DAPI in control and the indicated knockout HeLa cells. Insets show enlargements of the boxed areas. Right: Normalized line-scan intensity profiles of NuMA (green) and γ-tubulin (red) corresponding to the white dashed lines in left images. (E) 3D reconstruction of images shown in D. The length of the grid in the images is 3.65 µm. (F) Quantification of centrosome-to-centrosome distance and pole-to-pole distance (see Materials and methods for details) in control and indicated knockout HeLa cells. n = 40 cells for all conditions. (G) Left: Schematic representation of the distance between centrosome and pole (denoted by double-headed arrow), as measured from the center of γ-tubulin (γ-tub) spot (red) to the distal edge of NuMA spot (green). Right: Quantification of the distance between centrosome and pole in control and indicated knockout HeLa cells. n = 80 poles for all conditions. (H) Left: Schematic representation of outside and inside. The centers of two semicircular ROIs were selected to overlap with the visually determined center of the γ-tubulin spot (red). The chromosome- and cortex-directed ROIs were referred to as inside and outside, respectively. Right: Quantification of outside versus total intensity of γ-tubulin and NuMA in control and indicated knockout HeLa cells. n = 80 poles for all conditions. (I and J) Time-lapse images of PA-GFP-tubulin stripe in control or indicated knockout U2OS cells stably expressing PA-GFP–α-tubulin (I) and corresponding plots of the distance between PA-GFP stripe and spindle pole against time (J). (K) Quantification of minus-end depolymerization rate in control and indicated knockout U2OS cells. The minus-end depolymerization rate was derived from the plot slopes in J. Control, n = 33 cells; p80 knockout, n = 22; WDR62 knockout, n = 27. (L) Following cold treatment at 0°C for 5 min, the control and indicated knockout HeLa cells were fixed and immunostained for α-tubulin and DAPI. Maximum intensity projections of Z series with 30 stacks at 0.11-µm steps are shown. Arrows denote the curved spindle MTs. (M) Quantification of integrated intensity of spindle MTs in control and indicated knockout HeLa cells as shown in L. The intensity of each spindle was averaged from four middle stacks. n = 20 cells for all conditions. (N) Quantification of percentage of metaphase cells with normal or disorganized spindles in control and indicated knockout HeLa cells as shown in L. From left to right, n = 314, 280, 306 cells. Scale bars, 2 µm. Data represent mean ± SD. *, P < 0.05; ***, P < 0.001; two-tailed t test.
We previously showed that the rate of MT minus-end depolymerization at the spindle pole was dramatically reduced in p80 knockout cells, highlighting a role of katanin in the regulation of spindle dynamics (Jiang et al., 2017). To investigate whether WDR62 is also involved in this process, we knocked out WDR62 in U2OS cells stably coexpressing photoactivatable (PA)-GFP–α-tubulin and EB3-TagRFP (Fig. S1 J). Photoactivation experiments showed that the rate of minus-end depolymerization was strongly reduced in WDR62 knockout cells, to a similar extent as in p80 knockout cells (Fig. 2, I–K; and Video 1). Since the spindle pole or centrosome localization of KIF2A and KIF2C, which was previously reported to regulate spindle flux (Ganem et al., 2005), remained largely unaffected in WDR62 knockout HeLa cells (Fig. S1, K–Q), we conclude that WDR62 regulates spindle dynamics mainly by recruiting katanin to the spindle pole.
Video 1.
Reduced rate of MT minus-end depolymerization at the spindle pole in katanin p80 and WDR62 knockout cells. Photoactivation of PA-GFP–α-tubulin in control (left), p80 knockout (KO; middle), and WDR62 knockout (right) U2OS metaphase cells. Images were collected with a widefield fluorescent microscope at 2-s intervals. Video is sped up 30 times (15 frames/s). Time is shown in the format min:s.
To investigate whether the reduced rate of minus-end depolymerization impacts spindle stability, we treated cells with cold for 5 min. After cold treatment, ∼1.7-fold and ∼1.9-fold higher spindle MT intensity was observed in WDR62 knockout and p80 knockout HeLa cells, respectively, compared with control cells, demonstrating that spindle MTs become more stable in both knockout cells (Fig. 2, L and M). Even more strikingly, almost all knockout cells displayed disorganized spindle with curved MTs, which was not frequently observed in control cells (∼13%; Fig. 2, L and N). We assume that these abnormal, curved spindle MTs arise from buckling in response to the less efficient minus-end depolymerization. Collectively, we conclude that the WDR62–katanin complex regulates spindle organization and dynamics.
Biochemical characterization and mutational analysis of the WDR62–katanin complex
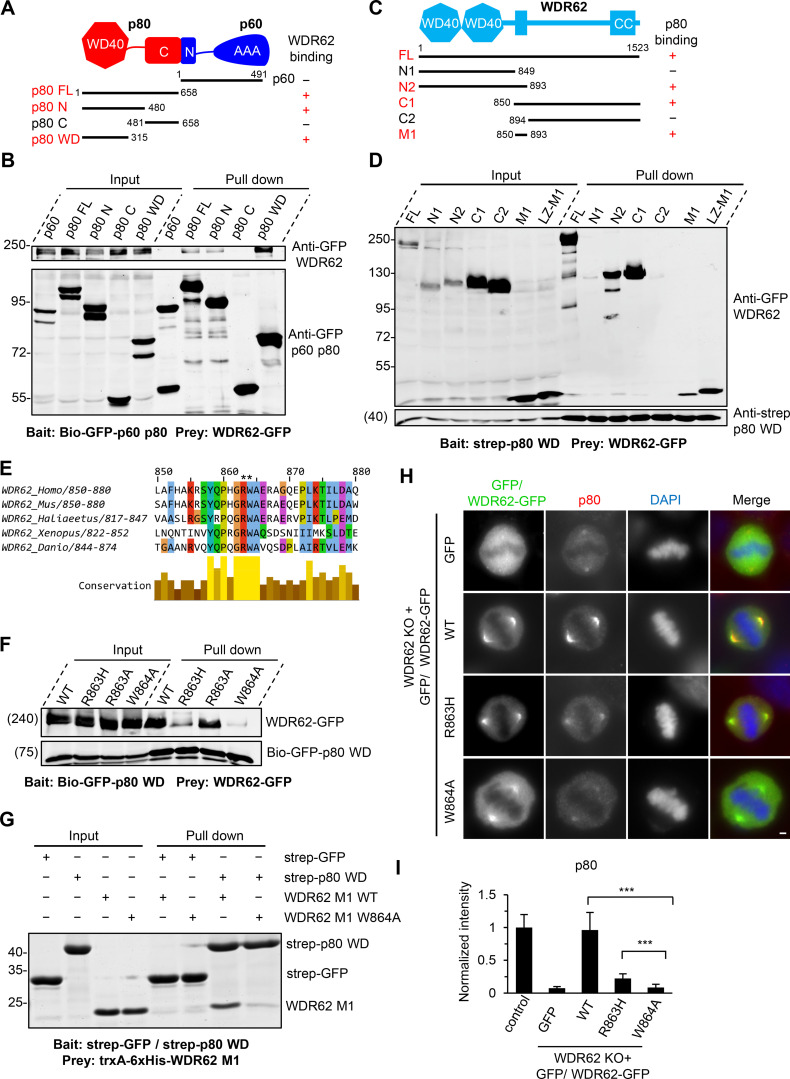
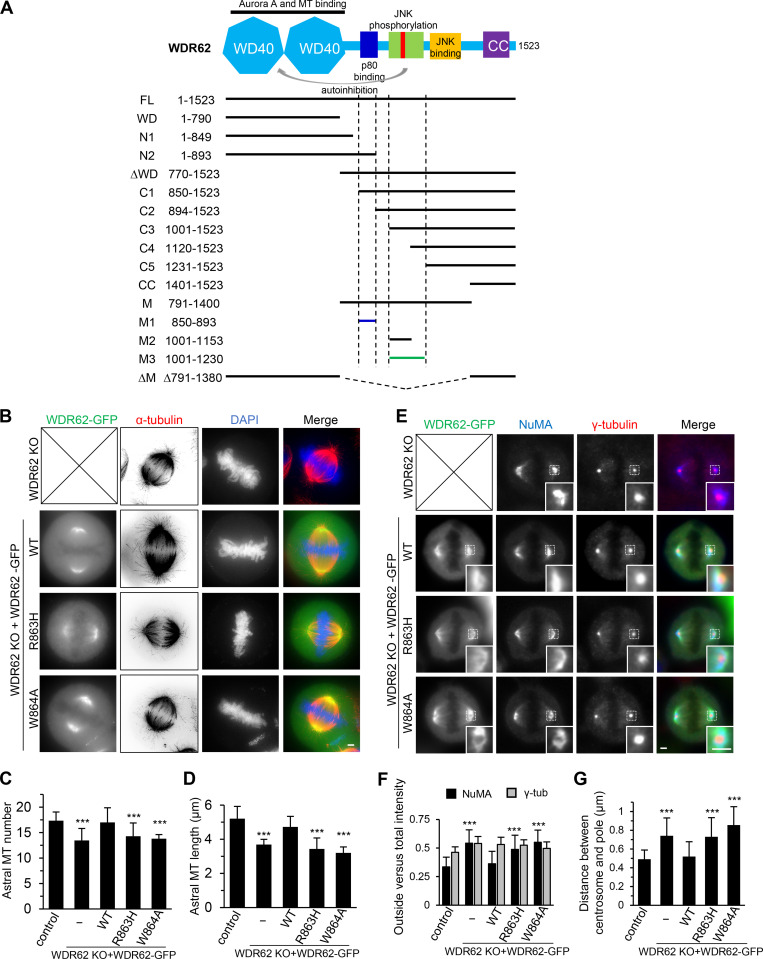
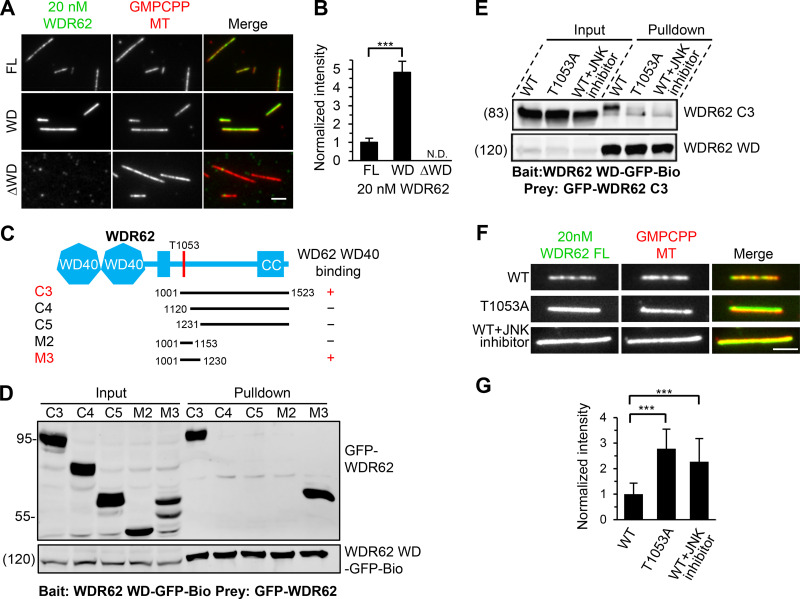
To explore the interaction between WDR62 and katanin in more detail, we performed pull-down assays. WDR62 interacts with the p80 subunit of katanin but not p60, and this interaction depends on the N-terminal WD40 domain (residues 1–315) of p80 (Fig. 3, A and B). We also mapped the p80-binding region of WDR62. WDR62 comprises an N-terminal WD40 domain, which is responsible for MT binding, followed by an unstructured region containing JNK binding and phosphorylation sites and a C-terminal coiled-coil domain required for its dimerization (Cohen-Katsenelson et al., 2011; Cohen-Katsenelson et al., 2013; Lim et al., 2015). By using deletion mutants of WDR62, we found that the interaction between p80 and WDR62 is mediated by a 44–aa stretch (residues 850–893) in the middle of WDR62 (fragment M1; Fig. 3, C and D; and Fig. S2 A), within which an MCPH-associated mutation R863H is positioned (Fig. 3 E; Poulton et al., 2014). Streptavidin pull-down assay showed that substitution of R863 by histidine (R863H) dramatically reduced the binding affinity of WDR62 for p80. Interestingly, substitution of R863 by alanine (R863A) had a relatively mild effect on the WDR62–p80 interaction; however, substitution of the neighboring residue W864 by alanine (W864A) almost completely abrogated this interaction, suggesting that the W864 residue is critical for this interaction while R863 also contributes to this interaction, albeit to a lesser extent than W864 (Fig. 3 F). Of note, both the R863 and W864 residues are evolutionarily conserved (Fig. 3 E). StrepTactin pull-down assay using purified proteins further validated a direct interaction between WDR62 and p80 and confirmed the importance of residue W864 (Fig. 3 G).To examine the effects of these mutations under physiological conditions, we introduced WT, R863H, and W864A versions of WDR62-GFP constructs into WDR62 knockout HeLa cells. All these constructs showed clear spindle pole localization. However, while the expression of WT WDR62-GFP could rescue the phenotypes caused by endogenous WDR62 knockout, including impaired accumulation of p80 at spindle poles, reduced number and length of astral MTs, and inward displacement of the centrosome relative to the spindle pole, expression of the R863H and W864A mutants failed to do so (Fig. 3, H and I; and Fig. S2, B–G), further validating the importance of the R863 and W864 residues. Thus, we conclude that WDR62 directly interacts with the p80 subunit of katanin, and the interface responsible for this interaction consists of the WD40 domain of p80 and residues 850–893 of WDR62.
Figure 3.
Biochemical characterization and mutational analysis of the WDR62–katanin complex.(A) Schematic overview of the domain organization of katanin p60/p80 heterodimer and the deletion mutants and summary of their interactions with WDR62. WD40, WD-40 repeat domain; C, C-terminus; N, N-terminus; AAA, AAA-ATPase domain. (B) Streptavidin pull-down assays with extracts of HEK293T cells expressing Bio-GFP–tagged p60 or p80 subunit or the indicated fragments (bait) together with WDR62-GFP (prey), analyzed by Western blotting with GFP antibody. (C) Schematic overview of the domain organization of WDR62 and the deletion mutants and summary of their interactions with p80. CC, coiled-coil domain. (D) StrepTactin pull-down assays with extracts of HEK293T cells expressing Strep-p80 WD40 domain (WD; bait) together with GFP-tagged WDR62 or its indicated fragments (prey) analyzed by Western blotting with indicated antibodies. LZ, leucine zipper from GCN4, was used as a dimerization domain. (E) Alignment of katanin-binding region in WDR62 from five vertebrate species. The conserved arginine and tryptophan residues are indicated with asterisks. (F) Streptavidin pull-down assays with extracts of HEK293T cells expressing the Bio-GFP-p80 WD (bait) together with GFP-tagged WT WDR62 or its indicated mutants (prey) analyzed by Western blotting with GFP antibody. (G) Coomassie blue–stained gel of StrepTactin pull down using Strep-p80 WD or Strep-GFP purified from HEK293T cells and immobilized on StrepTactin beads as the bait and WT WDR62 M1 fragment or its W864A mutant purified from E. coli as the prey. (H and I) Immunofluorescence staining and quantification of p80 intensities at spindle poles in WDR62 knockout (KO) HeLa cells transiently transfected with GFP-tagged WT WDR62 or its indicated mutants or control GFP vector. The values were normalized to the intensity of control HeLa cells. For all conditions, n = 74 spindle poles. Scale bar, 2 µm. Data represent mean ± SD. ∗∗∗, P < 0.001; two-tailed t test.
Figure S2.
Overview of WDR62 deletion mutants used in this study and the rescue of spindle defects by WT WDR62-GFP but not its katanin-binding deficient mutants.(A) Schematic illustration of the functional domains of WDR62 and summary of all the deletion mutants used in this study. WD40 domain (WD; light blue): MT binding and Aurora A binding; M1 (dark blue): p80 binding; M3: (green): autoinhibitory domain that interacts with WD40 domain. The horizontal lines were drawn to indicate the region spanned by the corresponding fragments (M1: dark blue; M3: green; others: black). The vertical dashed lines were drawn to indicate the boundaries of fragments M1 and M3. Note that JNK phosphorylation site T1053 (red) was located within the autoinhibitory M3 fragment. M, middle region; CC, coiled-coil. (B) Immunofluorescence staining of α-tubulin and DAPI in WDR62 knockout (KO) HeLa cells or the knockout cells transiently transfected with GFP-tagged WT WDR62 or its indicated mutants. Maximum intensity projections of Z series with 30 stacks at 0.11-µm steps are shown. (C and D) Quantification of the number and length of astral MTs shown in B. From left to right, n = 20, 20, 22, 20, and 20 spindle poles. (E) Immunofluorescence staining of NuMA and γ-tubulin in WDR62 knockout HeLa cells or the knockout cells transiently transfected with GFP-tagged WT WDR62 or its indicated mutants. Insets show enlargement of boxed areas. (F and G) Quantification of “outside” versus total intensity of γ-tubulin (γ-tub) and NuMA (F) and distance between the centrosome and the pole (G) shown in E. n = 50 spindle poles for all conditions in F; n = 48 spindle poles for all conditions in G. Scale bars, 2 µm. Data represent mean ± SD. ***, P < 0.001; two-tailed t test.
WDR62 shows a preference for extended MT lattices
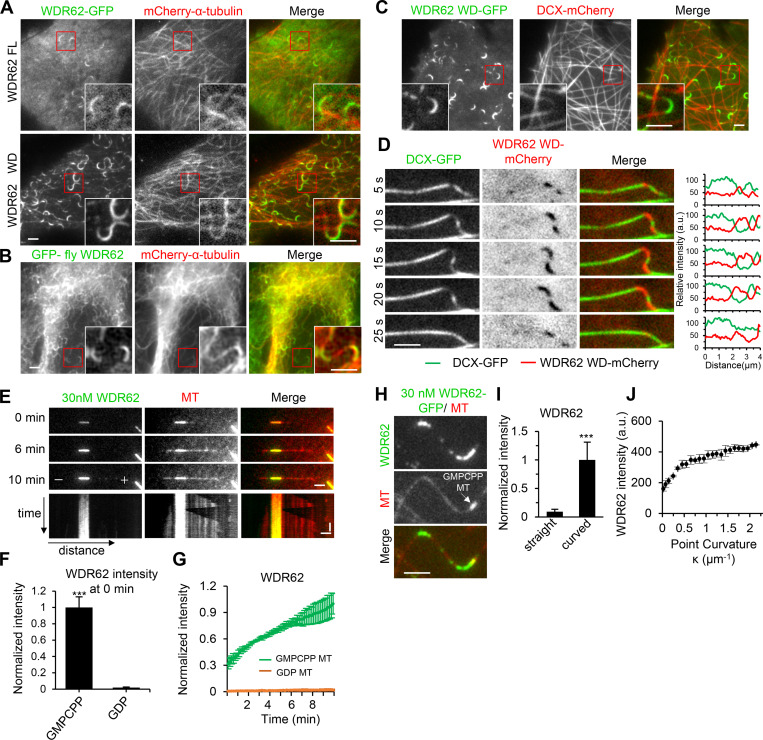
In previous studies, the localization of fluorescently tagged WDR62 was predominantly cytoplasmic in interphase cells; however, in the presence of paclitaxel, WDR62 binding evenly along MTs became readily apparent; in addition, its N-terminal fragment (1–841 aa) prominently decorated MTs during interphase (Lim et al., 2015). To better understand the MT-binding property of WDR62, we revisited its localization in MRC5 cells at low expression levels by applying total internal reflection fluorescence (TIRF) microscopy. Unexpectedly, we found that without paclitaxel treatment, WDR62-GFP preferentially accumulated at MT segments with high local curvature (Fig. 4 A and Video 2); moreover, its WD40 domain (1–790 aa; hereafter WD), compared with the full-length (FL) protein, displayed a higher affinity for curved regions (Fig. 4 A and Video 3). Interestingly, Drosophila melanogaster WDR62 tagged with GFP showed a similar preference for curved MTs in a human cell line (Fig. 4 B), suggesting that this activity is evolutionarily conserved.
Figure 4.
WDR62 preferentially binds to curved segments of dynamic MTs, as well as GMPCPP-stabilized MTs.(A) TIRF microscopy images of live MRC5 cells expressing GFP-tagged WDR62 FL or its WD40 domain (WD) together with mCherry–α tubulin. Insets show enlargements of the boxed areas. (B) Widefield fluorescence images of fixed MRC5 cell expressing GFP-fly WDR62 together with mCherry–α-tubulin. Insets show enlargements of the boxed areas. (C) TIRF microscopy images of live MRC5 cells expressing WDR62 WD-GFP together with DCX-mCherry. Insets show enlargements of the boxed areas. (D) Left: Time-lapse images showing the disappearance and reappearance of DCX-GFP and WDR62 WD-mCherry to mutually exclusive sites of a single MT, in response to MT buckling and straightening. Right: Corresponding fluorescence intensity profiles of WDR62 WD (red) and DCX (green) obtained from a line scan along the MT. (E) TIRF microscopy time-lapse images and kymograph showing gradual accumulation of WDR62-GFP (30 nM) on GMPCPP seed but not on the dynamic GDP lattice grown from the seed in a standard dynamic MT assay. (F) Quantification of WDR62-GFP intensity on GMPCPP seeds and GDP lattices at 0 min as shown in E. The values were normalized to the intensity on GMPCPP seeds. n = 23 MTs from three experiments. (G) Plot of WDR62-GFP intensity on GMPCPP seeds and GDP lattices against time. n = 23 MTs from three experiments. (H and I) TIRF microscopy images showing the preferential binding of WDR62-GFP (30 nM) to curved but not straight segments of dynamic GDP lattice in a flow-in assay (H) and quantification of the intensities of WDR62-GFP on straight and curved segments of GDP lattices (I). The arrow in H denotes the GMPCPP seed. The values were normalized to the intensity of curved segments. n = 87 MTs from three experiments. (J) Plot of WDR62-GFP intensity against the MT curvature κ, as shown in H. n = 31 MTs from three experiments. Horizontal scale bars, 2 µm; vertical scale bar, 2 min. Data represent mean ± SD. ***, P < 0.001; two-tailed t test.
Video 2.
WDR62-GFP preferentially accumulates at curved segments of dynamic MTs in cells. MRC5 cells were cotransfected with WDR62-GFP and mCherry–α-tubulin. Images were collected with a TIRF microscope in stream mode (2 frames/s). Video is sped up 10 times.
Video 3.
WDR62 WD-GFP displays strong affinity for curved segments of dynamic MTs in cells. MRC5 cells were cotransfected with WDR62 WD-GFP and mCherry–α-tubulin. Images were collected with a TIRF microscope in stream mode (2 frames/s). Video is sped up 10 times.
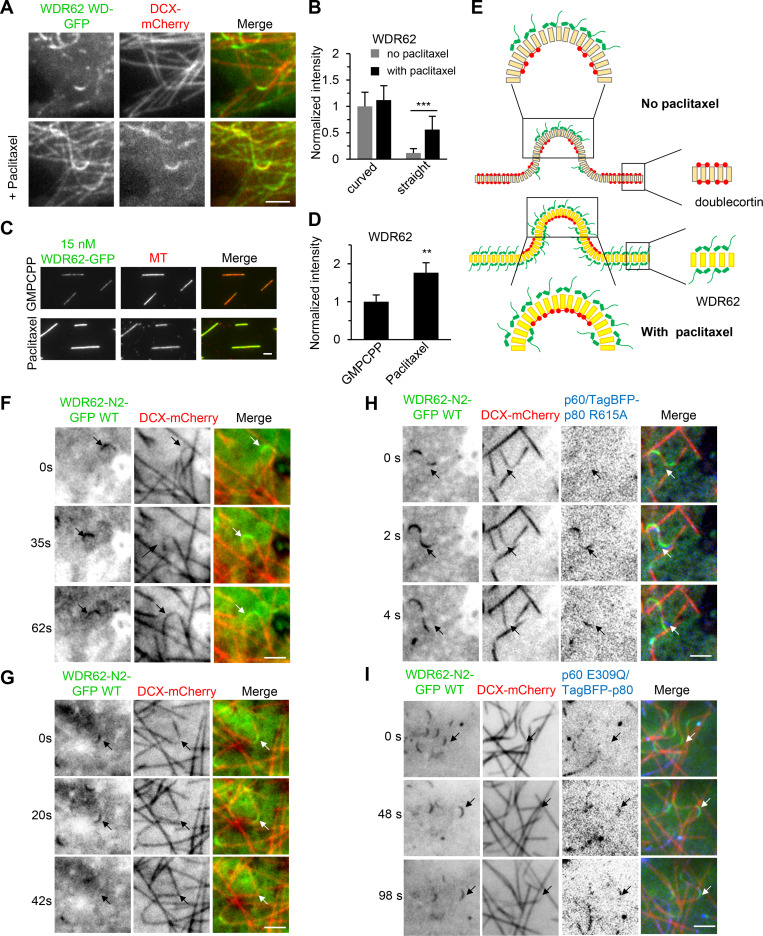
The MT geometry preference of WDR62 reminds us of Doublecortin (DCX), which associates with straight MTs in the absence of paclitaxel (Jean et al., 2012; Ettinger et al., 2016) but with curved MTs in the presence of paclitaxel (Bechstedt et al., 2014; Ettinger et al., 2016). Therefore, we examined the relative distributions of WDR62 and DCX in MRC5 cells. As WDR62 WD decorated curved MTs more evidently than its FL version, we cotransfected DCX with WDR62 WD, both fluorescently tagged. As expected, WDR62 WD and DCX displayed a mutually exclusive localization pattern along MTs, with WDR62 WD being most enriched at curved MT segments where the lowest DCX signal was observed (Fig. 4 C). Further time-lapse imaging of a single MT showed that WDR62 WD and DCX responded in an opposite and reversible manner to MT buckling and straightening (Fig. 4 D and Video 4). We then wondered whether paclitaxel can reverse the MT geometry preference of WDR62, as it did in the case of DCX (Ettinger et al., 2016). In contrast to its almost exclusive binding to curved regions of MTs in the absence of paclitaxel, WDR62 WD was distributed along both curved regions and straight parts in the presence of paclitaxel, although its intensity along the curved regions was still stronger than that along the straight parts (Fig. S3, A and B).
Video 4.
WDR62 WD-mCherry and DCX-GFP exhibit opposite responses to MT buckling and straightening. MRC5 cells were cotransfected with WDR62 WD-mCherry and DCX-GFP. Arrows indicate that WDR62 WD and DCX displayed a mutually exclusive localization pattern along MTs. Images were collected with a TIRF microscope in stream mode (2 frames/s). Video is sped up 10 times.
Figure S3.
Paclitaxel alters the MT lattice preference of WDR62, and katanin disassembles WDR62-decorated curved MT lattices.(A) TIRF microscopy images of live MRC5 cells expressing WDR62 WD-GFP together with DCX-mCherry before (top panels) or after (bottom panels) addition of 1 µM paclitaxel. (B) Quantification of the fluorescence intensities of WDR62 WD-GFP on straight and curved segments of dynamic MTs without or with paclitaxel treatment shown in A. The values were normalized to the intensity of curved segments without paclitaxel treatment. n = 30 MTs from three experiments for all conditions. (C and D) Images and quantification of the binding of WDR62-GFP (15 nM) to the GMPCPP- or paclitaxel-stabilized MTs in vitro. The values were normalized to the intensity of GMPCPP-stabilized MTs. From left to right, n = 196 and 163 MTs from three experiments. (E) Model of the recognition of extended MT lattice by WDR62. In the absence of paclitaxel, the outside protofilaments of the curved segments are in a more extended state compared with the rest of the MT lattices, explaining the preferential binding of WDR62 (green cartoon) to curved MTs both in cells and in vitro. In the presence of paclitaxel, except the inside curvature, which was occupied by DCX (red cartoon), the remainder of the MT lattices are all in an extended state, explaining why paclitaxel could change the MT geometry preference of WDR62. (F and G) Time-lapse images of live MRC5 cells expressing WDR62 N2-GFP together with DCX-mCherry. F represents the more frequently observed phenomenon (∼90%) that no breakage occurred (indicated with arrows) in WDR62 N2-GFP–decorated curved MTs, while G represents the less frequently observed phenomenon (∼10%) in which breakage occurred (indicated with arrows). (H and I) Time-lapse images of live MRC5 cells expressing p60/TagBFP-p80 R615A mutant or p60 E309Q/TagBFP-p80 mutant together with WDR62 N2-GFP and DCX-mCherry. Arrows indicate that p60/TagBFP-p80 R615A (H), but not p60 E309Q/TagBFP-p80 (I), could disassemble curved MT lattices, although both katanin mutants were recruited there by WDR62 N2-GFP. DCX-mCherry was used to distinguish between straight and curved MT lattices. Scale bars, 2 µm. Data represent mean ± SD. **, P < 0.01; ***, P < 0.001; two-tailed t test.
We then set out to investigate MT-binding activity of WDR62 in vitro. To this end, recombinant FL WDR62 was purified from HEK293T cells through one-step affinity purification with StrepTactin beads (Fig. S5 G). We started by performing an in vitro dynamic assay according to a standard procedure described previously (Mohan et al., 2013). We noticed that WDR62 preferentially decorated the GMPCPP seeds and its intensity along seeds increased progressively over time; however, accumulation of WDR62 was hardly observed along the GDP-MTs grown from the seeds (Fig. 4, E–G). Since WDR62 showed a preference for curved MTs in cells, we next sought to determine whether this phenomenon could be recapitulated in vitro. Therefore, we used an alternative protocol aiming at producing curved MTs in vitro (see Materials and methods for details). WDR62 strongly accumulated along the curved regions but not straight parts of GDP-MTs (Fig. 4, H and I). Moreover, its intensity along the curved region increased with increasing local curvature (Fig. 4 J). We next examined the effect of paclitaxel on the WDR62-MT association in vitro. We found that WDR62 displayed strong binding to paclitaxel-stabilized MTs, and the WDR62 intensity in this case was even approximately twofold enhanced compared with that observed with GMPCPP-stabilized MTs (Fig. S3, C and D).
Figure S5.
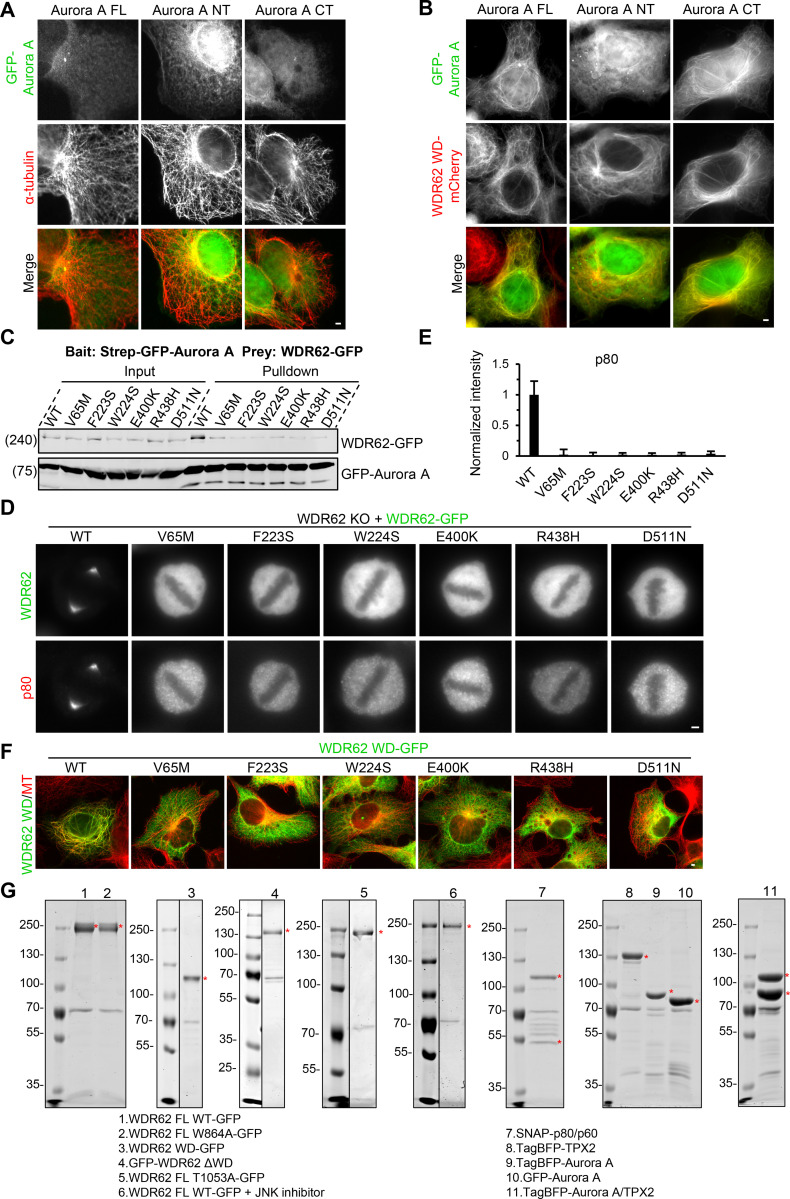
MCPH-associated mutations within WD40 domain of WDR62 disrupt its interactions with both MTs and Aurora A.(A) Immunofluorescence staining of α-tubulin in MRC5 cells transiently expressing GFP-tagged Aurora A FL, N-terminal part (NT), or C-terminal kinase domain (CT). All these Aurora A constructs by themselves failed to localize to MTs. (B) Images of fixed MRC5 cells transiently expressing WDR62 WD-mCherry together with GFP-tagged Aurora A FL, NT, or CT. WDR62 WD-mCherry can recruit GFP-tagged Aurora A FL and CT, but not NT, to MTs. (C) StrepTactin pull-down assays with extracts of HEK293T cells expressing Strep-GFP-Aurora A (bait) together with GFP-tagged WT WDR62 or its indicated MCPH-associated mutants (prey). (D and E) Immunofluorescence staining and quantification of katanin p80 intensities at spindle poles in WDR62 knockout (KO) HeLa cells transiently transfected with GFP-tagged WT WDR62 or its indicated MCPH-associated mutants. For all conditions, n = 42 spindle poles. (F) Immunofluorescence staining of α-tubulins in MRC5 cells transiently transfected with GFP-tagged WT WDR62 or its indicated MCPH-associated mutants. (G) Purified proteins used for in vitro assays. The bands corresponding to indicated proteins are marked with red asterisks on Coomassie blue–stained gels. Note that molecular chaperone HSP70 (∼70 KD) was copurified with all indicated proteins. Scale bars, 2 µm. Data represent mean ± SD.
Taken together, our data show that WDR62 preferentially binds to GMPCPP- and paclitaxel-stabilized MTs, both of which have a more extended lattice structure compared with GDP-MTs (Alushin et al., 2014), as well as curved dynamic GDP-MTs, the outside protofilaments of which, in theory, are also in an extended state (Ettinger et al., 2016). We therefore propose a model in which WDR62 recognizes the extended MT lattice (Fig. S3 E). Since DCX has been proposed to recognize the compacted GDP-MT lattice (Ettinger et al., 2016), our model can also explain the series of opposite behaviors of WDR62 and DCX described above.
JNK phosphorylation–mediated autoinhibition regulates MT-binding affinity of WDR62
As shown in Fig. 4 A, the MT-binding affinity of WDR62 WD was much stronger than that of the FL protein in cells. Consistently, our in vitro assays using purified proteins showed that the affinity of WDR62 WD for GMPCPP-stabilized MTs increased fourfold to fivefold compared with the FL protein, whereas the deletion mutant lacking WD (ΔWD) completely lost such affinity (Fig. 5, A and B; and Fig. S5 G). We reasoned that the WDR62 molecule may exist in a folded conformation, which allows its other regions to directly contact WD, thereby potentially masking some of the MT-binding sites on WD. To test this possibility, we performed a streptavidin pull-down assay, using WD as bait and middle or C-terminal fragments as prey. Both fragments C3 (residues 1001–1523) and M3 (residues 1001–1230) could be pulled down by WD, demonstrating that intramolecular interaction in WDR62 indeed exists. Sequence analysis of fragment M3 revealed that JNK phosphorylation site T1053 is located within this minimal WD-binding region (Fig. 5, C and D; and Fig. S2 A). Whereas JNK phosphorylation was previously reported to negatively modulate association of WDR62 with MTs (Lim et al., 2015), the precise mechanism remains undetermined. Based on the published results and our observations described here, we further hypothesized that JNK phosphorylation within the M3 region might induce its intramolecular interaction with WD, thus leading to an autoinhibited conformation. To support this, our pull-down assay showed that introducing of T1053A substitution into fragment C3 or the presence of JNK inhibitor can abrogate the interaction between WD and fragment C3 (Fig. 5 E). Consistently, our in vitro experiments showed that introduction of T1053A substitution into the recombinant FL WDR62 led to a marked increase in its MT-binding affinity compared with the WT version and that a similar increase was observed when the WT WDR62 was expressed and purified in the presence of JNK inhibitor (Fig. 5, F and G; and Fig. S5 G). We conclude that JNK phosphorylation can mediate autoinhibition of WDR62, thereby negatively modulating WDR62 association with MTs.
Figure 5.
The MT-binding affinity of WDR62 is autoinhibited through JNK phosphorylation–induced intramolecular interaction.(A and B) Images and quantification of the binding of 20 nM GFP-tagged WDR62 FL or its indicated truncations (green) to GMPCPP-stabilized MTs (red). The values were normalized to the intensity of the WDR62 FL-GFP. N.D., not detected. For all conditions, n = 60 MTs from two experiments. (C) Schematic overview of the domain organization of WDR62 and the deletion mutants and summary of their interactions with WD40 domain. CC, coiled-coil. (D) Streptavidin pull-down assays with extracts of HEK293T cells expressing WDR62-WD-GFP-Bio (bait) together with the indicated GFP-tagged WDR62 truncations (prey) analyzed by Western blotting with GFP antibody. (E) Streptavidin pull-down assays with extracts of HEK293T cells expressing WDR62-WD-GFP-Bio (bait) together with GFP-tagged WT WDR62 C3 fragment with or without JNK inhibitor treatment or its T1053A mutant (prey) analyzed by Western blotting with GFP antibody. (F and G) Images and quantification of the binding of 20 nM GFP-tagged FL WDR62 WT with or without JNK inhibitor treatment or T1053A mutant (green) to GMPCPP-stabilized MTs (red). The values were normalized to the intensity of the WT WDR62-GFP without JNK inhibitor treatment. From left to right, n = 150, 150, and 124 MTs from three experiments. Scale bars, 2 µm. Data represent mean ± SD. ∗∗∗, P < 0.001; two-tailed t test.
WDR62 recruits katanin to sever curved MTs in cells and GMPCPP-stabilized MTs in vitro
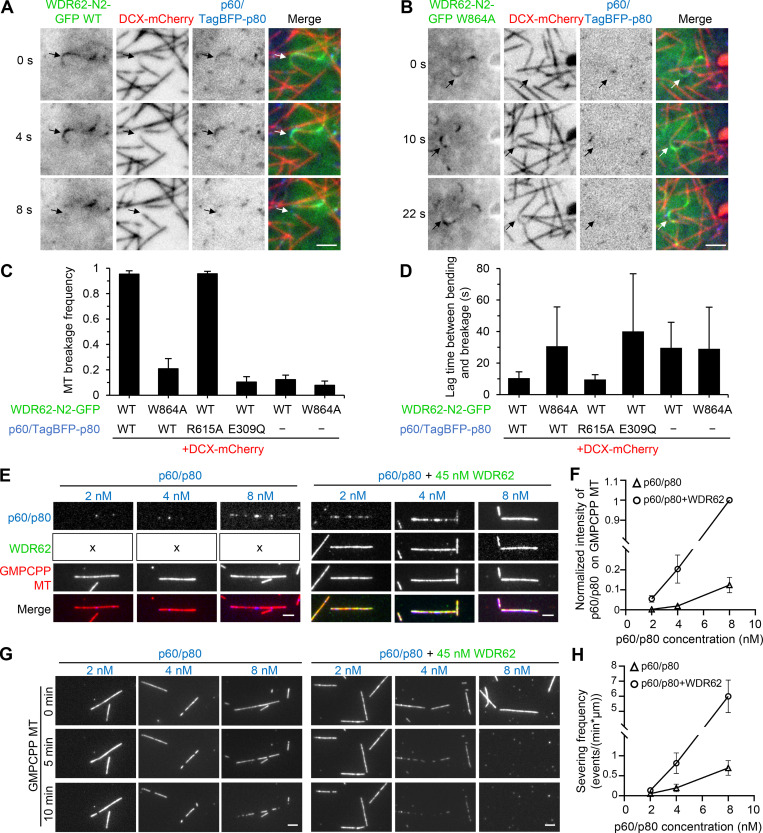
We then set out to investigate the combined activities of WDR62 and katanin. To better visualize WDR62 behavior in MRC5 cells, we used WDR62 fragment N2 (WDR62-N2-GFP), which contains both WD and p80-binding region (Fig. 3 C and Fig. S2 A). Live-cell imaging together with quantification analyses showed that in the presence of WT WDR62-N2-GFP or the katanin binding–deficient mutant WDR62-N2-W864A-GFP alone, ∼10% of curved MTs could break, most likely due to mechanical stress, and the average lag time between appearance of bending and breakage was ∼30 s (Fig. 6, C and D; and Fig. S3, F and G). When WDR62-N2-GFP was cotransfected with p60/TagBFP-p80 or p60/TagBFP-p80 R615A, which is deficient in MT end binding and bending (Jiang et al., 2017), both versions of katanin could be recruited to curved MTs, 95% of which subsequently broke, and the average lag time between appearance of bending and breakage was ∼10 s (Fig. 6, A, C, and D; Fig. S3 H; and Video 5), suggesting that katanin-mediated severing was much faster and more efficient than the mechanical break caused by WDR62 N2 alone. As expected, when WDR62-N2-W864A-GFP was cotransfected with p60/TagBFP-p80 or when WDR62-N2-GFP was cotransfected with p60 E309Q/TagBFP-p80, which is catalytically inactive (Eckert et al., 2012; Shin et al., 2019), curved MT breakage occurred to a similar extent as with WDR62 N2 alone (Fig. 6, B–D; and Fig. S3 I). Collectively, our data showed that in cells, WDR62 can recruit katanin to sever curved MTs, but the end-binding/bending activity of katanin is dispensable for this process.
Figure 6.
WDR62 recruits katanin to sever curved MTs in cells and GMPCPP-MTs in vitro.(A and B) Time-lapse images of live MRC5 cells expressing GFP-tagged WT WDR62 N2 fragment (A) or its W864A mutant (B) together with DCX-mCherry and p60/TagBFP-p80. Arrows showing that p60/TagBFP-p80 was recruited to curved MT lattices by WDR62 N2 WT-GFP (A), but not by WDR62 N2 W864A-GFP (B), and that breakage occurred in the former but not the latter case. DCX-mCherry was used to distinguish between straight and curved MT lattices. (C) Quantification of MT breakage frequency in MRC5 cells expressing indicated constructs. n = 3 experiments; 90–120 curved MTs were measured. (D) Quantification of lag time between appearance of bending and breakage in MRC5 cells expressing indicated constructs. n = 90–100 breakage events for WDR62 N2 WT-GFP cotransfected with p60/TagBFP-p80 or p60/TagBFP-p80 R615A; n = 10–30 for other conditions. (E and F) Images and quantification of p60/SNAP-p80 (blue) intensity on GMPCPP-stabilized MTs (red) at various concentrations (2 nM, 4 nM, and 8 nM) in the absence (indicated with "x" in the left panels) or presence of 45 nM WDR62-GFP (green). The values were normalized to the intensity of 8 nM p60/SNAP-p80 in the presence of 45 nM WDR62-GFP. n = 40–60 MTs from three experiments. (G and H) Images and quantification of MT-severing frequency of p60/SNAP-p80 (2 nM, 4 nM, and 8 nM) in the absence or presence of 45 nM WDR62-GFP. n = 3 experiments; 40–60 MTs were measured. Scale bars, 2 µm. Data represent mean ± SD.
Video 5.
WT WDR62 N2-GFP recruits p60/TagBFP-p80 to sever curved MTs in cells. MRC5 cells were cotransfected with WT WDR62 N2-GFP, DCX-mCherry, and p60/TagBFP-p80. Arrows indicate that p60/TagBFP-p80 was recruited to curved MT lattices by WDR62 N2 WT-GFP and that breakage occurred. Images were collected with a TIRF microscope at 2-s intervals. Video is sped up 10 times (5 frames/s).
We then asked whether WDR62 can recruit katanin to sever MTs in vitro. The severing activity of FL katanin p60/p80 purified from HEK293T cells (Fig. S5 G) was investigated over a range of concentrations from 2–8 nM in the absence or presence of a fixed concentration of WDR62 (45 nM). Addition of WDR62 led to an increased accumulation of p60/p80 along GMPCPP-stabilized MTs at all tested concentrations (Fig. 6, E and F). Consistently, the severing frequency of p60/p80 also increased, with the increase being more prominent at 4 nM and 8 nM (fourfold for 4 nM, 8.5-fold for 8 nM; Fig. 6, G and H). WDR62 can thus enhance the severing activity of katanin by recruiting it to GMPCPP-stabilized MTs.
Taken together, our data demonstrate that WDR62 can recruit katanin to curved MTs in cells and to GMPCPP-stabilized MTs in vitro, suggesting that WDR62 by itself specifically targets katanin to sever extended MT lattices.
TPX2–Aurora A–WDR62–katanin pathway exists in cells
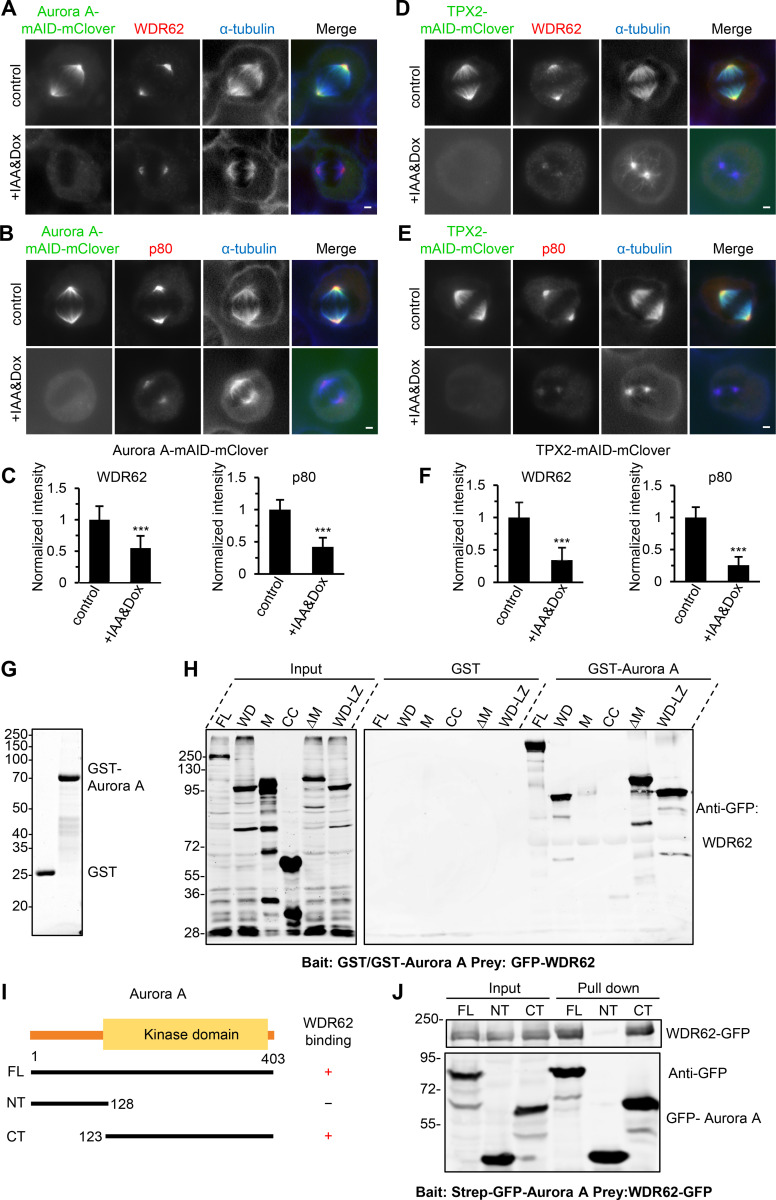
Although WDR62 displayed an intrinsic affinity for extended MT lattices, MT lattices around spindle poles, in most cases, are unlikely to be in this special conformational state. Thus, the prominent spindle pole localization of WDR62 could not be attributed to its own MT-binding activity but instead should be determined by other recruiting factors. Previously, it was reported that Aurora A and its upstream coactivator TPX2 can recruit WDR62 to the spindle pole (Lim et al., 2015; Lim et al., 2016). Together with our data showing that WDR62 can recruit katanin to the spindle pole, it is reasonable to predict that katanin acts in the same pathway as WDR62, Aurora A, and TPX2. To test this prediction, we performed conditional knockdown of proteins using the AID system (Nishimura et al., 2009). Upon treatment with Indole-3-acetic acid (IAA) for 12 h, TPX2 and Aurora A could be knocked down by 93% and 86%, respectively, without significantly reducing the expression of each other or WDR62 and katanin (Fig. S4, A–D). Depletion of TPX2 and Aurora A resulted in mitotic arrest and severe spindle defects, including monopolar spindle and fragmented spindle, as well as prometaphase-like spindle (TPX2 depletion only) or small bipolar spindle (Aurora A depletion only; Fig. S4, E–J), similar to those described previously (Garrett et al., 2002; Gruss et al., 2002; Kufer et al., 2002; Bird and Hyman, 2008). To examine the effect of depletion of TPX2 or Aurora A on the spindle pole localization of WDR62 and katanin, we only scored cells with two spindle poles, and to enrich the population of these cells, we treated cells with IAA for a shorter period (6 h). As previously described, TPX2 is required for the spindle localization of Aurora A (Fig. S4, K and L). And depletion of TPX2 or Aurora A resulted in the expected impaired accumulation of WDR62 and katanin at the spindle pole, while knockout of WDR62 or katanin had little impact on the spindle pole localization of TPX2 and Aurora A (Fig. 7, A–F; and Fig. S4, M–P). Based on the sequential recruitment observed in previous works and in this study, we conclude that these four proteins interact and assemble at the spindle pole in a hierarchy, with TPX2 at the apex and Aurora A, WDR62, and katanin at sequentially lower positions.
Figure S4.
Characterization of TPX2- or Aurora A–mAID-mClover knock-in cell lines.(A) Schematic representation of the strategy to construct conditional AID mutant of TPX2 or Aurora A via CRISPR/Cas9 as well as primer sets for genotyping and the expected PCR products. Tet-on 3G, Tet-On 3G transactivator; pPGK, Phosphoglycerate kinase 1 promoter; pTRE3GS, TRE3GS promoter; KI, knock-in; Puro, puromycin resistance gene; Hygro, hygromycin resistance gene. Parental cell lines were first generated by introducing 3xHA-OsTIR1 at the AAVS1 locus. Subsequently, the mAID-mClover cassette was introduced into parental cells after the last codon of TPX2 or Aurora A. (B and C) The homozygous TPX2- or Aurora A (AurA)–mAID-mClover knock-in HeLa cell lines were confirmed by Western blotting with indicated antibodies (B) and PCR genotyping (C). (D) Western blotting analysis of indicated protein levels in TPX2- or Aurora A–mAID-mClover knock-in HeLa cells without or with doxycycline (Dox) and IAA treatment. For all conditions, cells were synchronized in mitosis with STLC. (E and F) Images showing indicated spindle defects in TPX2- or Aurora A–mAID-mClover knock-in HeLa cells treated with doxycycline and IAA. (G) Quantification of mitotic index in TPX2- or Aurora A–mAID-mClover knock-in HeLa cells without or with doxycycline and IAA treatment, n = 3 experiments. For TPX2-mAID-mClover, control cells, 1,113 cells; doxycycline and IAA treatment, 838 cells. For Aurora A–mAID-mClover, control, 1,126 cells; doxycycline and IAA treatment, 1,142 cells. (H and I) Percentage of cells with indicated spindle phenotypes in TPX2- or Aurora A–mAID-mClover knock-in HeLa cells in prometaphase or metaphase without or with doxycycline and IAA treatment. For TPX2-mAID-mClover, control, n = 221 cells; doxycycline and IAA treatment, n = 244 cells. For Aurora A–mAID-mClover, control, n = 231 cells; doxycycline and IAA treatment, n = 220 cells. (J) Quantification of the length of bipolar spindle in Aurora A–mAID-mClover knock-in HeLa cells without or with doxycycline and IAA treatment. For both conditions, n = 40 cells. (K and L) Immunofluorescence staining and quantification of Aurora A intensity at spindle poles in TPX2-mAID-mClover knock-in HeLa cells without or with doxycycline and IAA treatment. For both conditions, n = 100 spindle poles. (M–P) Immunofluorescence staining and quantification of Aurora A and TPX2 intensities at spindle poles in control or the indicated knockout (KO) HeLa cells. For all conditions, n = 100 spindle poles. Scale bars, 2 µm. Data represent mean ± SD. ***, P < 0.001; two-tailed t test.
Figure 7.
Validation of a TPX2–Aurora A–WDR62–katanin axis in cells.(A–C) Immunofluorescence staining and quantification of WDR62 and katanin p80 intensities at spindle poles in Aurora A–mAID-mClover knock-in HeLa cells without or with doxycycline (Dox) and IAA treatment. For WDR62 intensity, control, n = 102 spindle poles; doxycycline and IAA treatment, n = 108; for p80 intensity, both control and doxycycline and IAA treatment, n = 104. (D–F) Immunofluorescence staining and quantification of WDR62 and katanin p80 intensities at spindle poles in TPX2-mAID-mClover knock-in HeLa cells without or with doxycycline and IAA treatment. For all conditions, n = 100 spindle poles. (G) Coomassie blue–stained gel with GST and GST–Aurora A purified from E. coli and used for the GST pull-down assays in H. (H) GST or GST–Aurora A pull-down assays with extracts of HEK293T cells expressing GFP-tagged WDR62 FL or its indicated fragments analyzed by Western blotting with GFP antibody. M, middle region; CC, coiled-coil; LZ, leucine zipper. (I) Schematic overview of the domain organization of Aurora A and the deletion mutants and summary of their interactions with WDR62. (J) StrepTactin pull-down assays with extracts of HEK293T cells expressing Strep-GFP–tagged Aurora A FL, N-terminal part (NT), or C-terminal kinase domain (CT; bait) together with WDR62-GFP (prey) analyzed by Western blotting with GFP antibody. Scale bars, 2 µm. Data represent mean ± SD. ***, P < 0.001; two-tailed t test.
As controversy exists over the molecular details of the Aurora A–WDR62 interaction (Chen et al., 2014; Lim et al., 2016), we next investigated this issue by performing pull-down assays. We found that WDR62 interacts with Aurora A through WD, which is also responsible for its association with MTs (Fig. 7, G and H; and Fig. S2 A). We also mapped the WDR62-binding region of Aurora A to its C-terminal kinase domain (Fig. 7, I and J). Consistently, we observed by fluorescent microscopy that in MRC5 cells WDR62 WD-mCherry can recruit GFP-tagged Aurora A FL and its C-terminal kinase domain, but not its N-terminal part, to MTs (Fig. S5, A and B). In addition, the fact that WDR62 WD can bind to MT and Aurora A simultaneously indicates the presence of distinct binding sites for MT and Aurora A within WD. Previous study had shown that two MCPH-associated missense mutations (V65M, R438H) within WDR62 WD can impair the spindle pole localization of WDR62 and perturb the WDR62–Aurora A interaction (Lim et al., 2015; Lim et al., 2016). We then extended this analysis and examined most MCPH-associated missense mutations, including V65M, F223S, W224S, E400K, R438H, and D511N, located within WD (Shohayeb et al., 2018). We found that ectopically expressed FL WDR62 harboring these individual mutations failed to interact with Aurora A and displayed mislocalization in mitosis and a corresponding failure to rescue the spindle pole localization of p80 in WDR62 knockout HeLa cells (Fig. S5, C–E). Moreover, all these mutations perturbed the MT-binding activity of WDR62 WD in interphase MRC5 cells (Fig. S5 F). Therefore, interference with WDR62 interactions with Aurora A and MT might be general consequences of these MCPH-associated mutations, most likely caused by the misfolding of WDR62 WD.
TPX2–Aurora A–WDR62–katanin pathway promotes disassembly of GDP-MTs in vitro
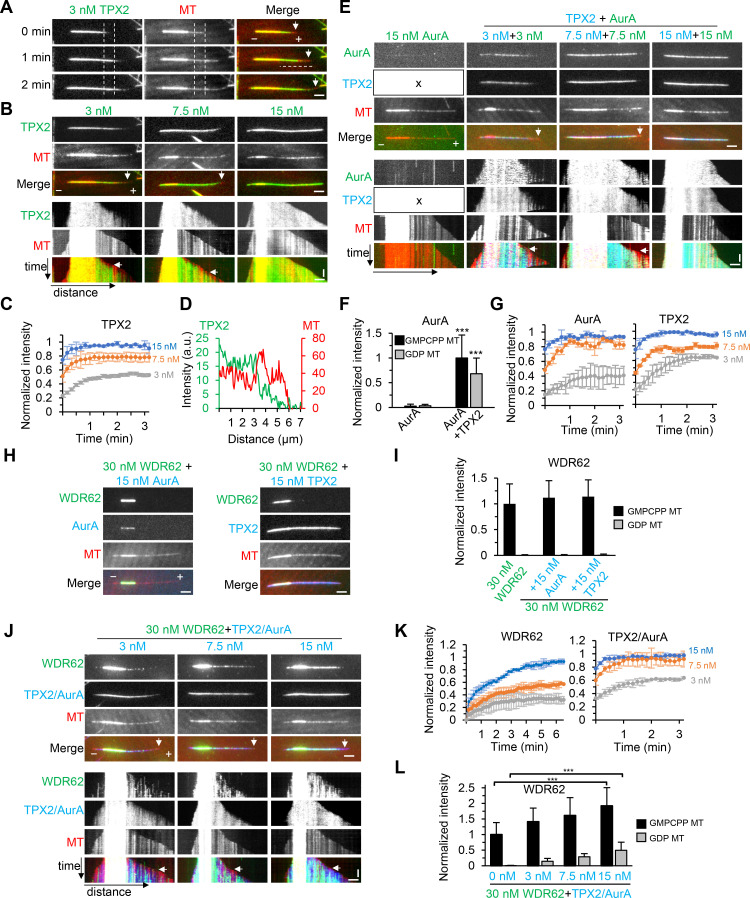
As mentioned above, WDR62 alone can only recruit katanin to sever GMPCPP-stabilized MTs in vitro. We then asked whether TPX2/Aurora A can recruit WDR62 to straight GDP-MTs, which in turn targets katanin and promotes katanin-mediated severing of GDP-MTs. To this end, recombinant TPX2 and Aurora A were either purified individually or copurified from HEK293T cells through one-step affinity purification with StrepTactin beads (Fig. S5 G). Varying concentrations of these proteins were subsequently applied into the in vitro dynamic MT assays. In our assay conditions, besides seeds binding, TPX2 also localized to GDP-MTs and accumulated progressively over time until reaching a plateau, leading to biased enrichment of TPX2 toward older regions of GDP-MTs, with the tendency being more prominent at lower concentrations (3 nM and 7.5 nM; Fig. 8, A–D; and Fig. 8, A and B, arrows, denoting no binding of TPX2 to newly generated segments of GDP-MTs). Similar observations were described for TPX2 in Xenopus laevis egg extracts (Thawani et al., 2019). Although MT binding was hardly observed for Aurora A alone, in the presence of TPX2, which was used at the same concentrations as Aurora A to form a 1:1 complex, Aurora A was strongly recruited and exhibited a similar distribution pattern along MTs as TPX2 (Fig. 8, E–G).
Figure 8.
TPX2/Aurora A complex recruits WDR62 to GDP-MTs in vitro. (A) Time-lapse images showing progressive accumulation of 3 nM TagBFP-TPX2 (green) on GDP-MT over time (compare the TPX2 intensity between two vertical dashed lines at indicated time points), leading to biased deposition of TPX2 on older lattice segment near the seed and no binding to newly generated segment of GDP-MT (arrows). The distance between two vertical dashed lines is 2 µm, and the position of the right dashed line is MT plus end at 0 min. (B) Images and corresponding kymographs showing MT binding of 3 nM, 7.5 nM, and 15 nM TagBFP-TPX2 (green). Vertical arrows in images and horizontal arrows in kymographs indicate the absence of TPX2 (3 nM and 7.5 nM) from newly generated segment of GDP-MT. (C) Plots of average intensity of TagBFP-TPX2 at indicated concentrations on a 2-µm-long segment of GDP-MT as in A (between two vertical dashed lines) against time. The values were normalized to the maximum intensity of 15 nM TPX2. n = 10–20 MTs from two experiments. (D) Plots of intensities of TagBFP-TPX2 (green) and MT (red) against distance (denoted by the horizontal dashed line at 1 min in A), showing stronger TPX2 intensity on older GDP lattice. (E) Images and corresponding kymographs of GFP–Aurora A (AurA) alone (green) or together with TagBFP-TPX2 (blue) at indicated concentrations. In the presence of TPX2, Aurora A was recruited to MTs and colocalized with TPX2. The "x" in left panels indicates that TPX2 was not included in the assay. Vertical arrows in images and horizontal arrows in kymographs indicate the absence of TPX2 and Aurora A (3 nM and 7.5 nM) from newly generated segment of GDP-MT. (F) Quantification of intensities of 15 nM GFP–Aurora A on GMPCPP seeds and GDP MTs in the absence or presence of 15 nM TagBFP-TPX2 after 5 min of GDP-MT growth from seed. n = 21 or 22 MTs from two experiments. (G) Plots of average TagBFP-TPX2 or GFP–Aurora A intensity on a 2-µm-long segment of GDP-MT at indicated concentrations against time as in C. The values were normalized to the maximum intensity in the condition of TPX2 and Aurora A both at 15 nM. n = 10–20 MTs from two experiments. (H) Images of MTs grown in the presence of 30 nM WDR62-GFP (green) together with 15 nM TagBFP–Aurora A (blue) or 15 nM TagBFP-TPX2 (blue). Note that Aurora A can weakly localize to seeds in the presence of WDR62 (compare E and H). (I) Quantification of intensities of 30 nM WDR62-GFP on the GMPCPP seeds and GDP MTs in the absence or presence of 15 nM TagBFP–Aurora A or 15 nM TagBFP-TPX2 after 5 min of GDP-MT growth from seed. n = 20–30 MTs from two experiments. (J) Images and corresponding kymographs of MTs grown in the presence of WDR62-GFP (green) and TPX2/TagBFP–Aurora A complex (blue) at indicated concentrations. Vertical arrows in images and horizontal arrows in kymographs indicate the absence of WDR62 from newly generated segment of GDP-MT at all TPX2/Aurora A concentrations. (K) Plots of average WDR62-GFP or TPX2/TagBFP–Aurora A intensity on a 2-µm-long segment of GDP-MT at indicated concentrations against time as in C. The values were normalized to the maximum intensity in the condition of 30 nM WDR62 with 15 nM TPX2/Aurora A. n = 10–16 MTs from two experiments. (L) Quantification of intensities of 30 nM WDR62-GFP on GMPCPP seeds and GDP MTs in the absence or presence of TPX2/TagBFP–Aurora A at indicated concentrations after 5 min of GDP-MT growth from seed. The values were normalized to the intensity of WDR62 alone on GMPCPP seeds. n = 17–30 MTs from two experiments. Horizontal scale bar, 2 µm; vertical scale bar, 1 min. Data represent mean ± SD. ***, P < 0.001; two-tailed t test.
We next examined the effects of TPX2, Aurora A, or the copurified TPX2/Aurora A complex on WDR62 behavior in vitro. As expected, the addition of either TPX2 or Aurora A failed to recruit WDR62 to GDP-MTs (Fig. 8, H and I). By contrast, addition of increasing concentrations of TPX2/Aurora A complex caused increased recruitment of WDR62 to GDP-MTs. Consequently, WDR62 displayed a similar biased distribution pattern toward older regions of GDP-MTs as TPX2/Aurora A complex. However, WDR62 and TPX2/Aurora A showed different kinetics. Especially at 15 nM TPX2/Aurora A, at which concentration TPX2/Aurora A quickly reached peak intensity within 1 min, WDR62 intensity persistently increased during the 6-min imaging window (Fig. 8, J–L). Therefore, WDR62 displayed stronger “older GDP-MT region” bias than TPX2. Of note, although WDR62 itself displayed strong affinity for GMPCPP seeds, its intensity along seeds also increased with increasing TPX2/Aurora A concentration (Fig. 8 L).
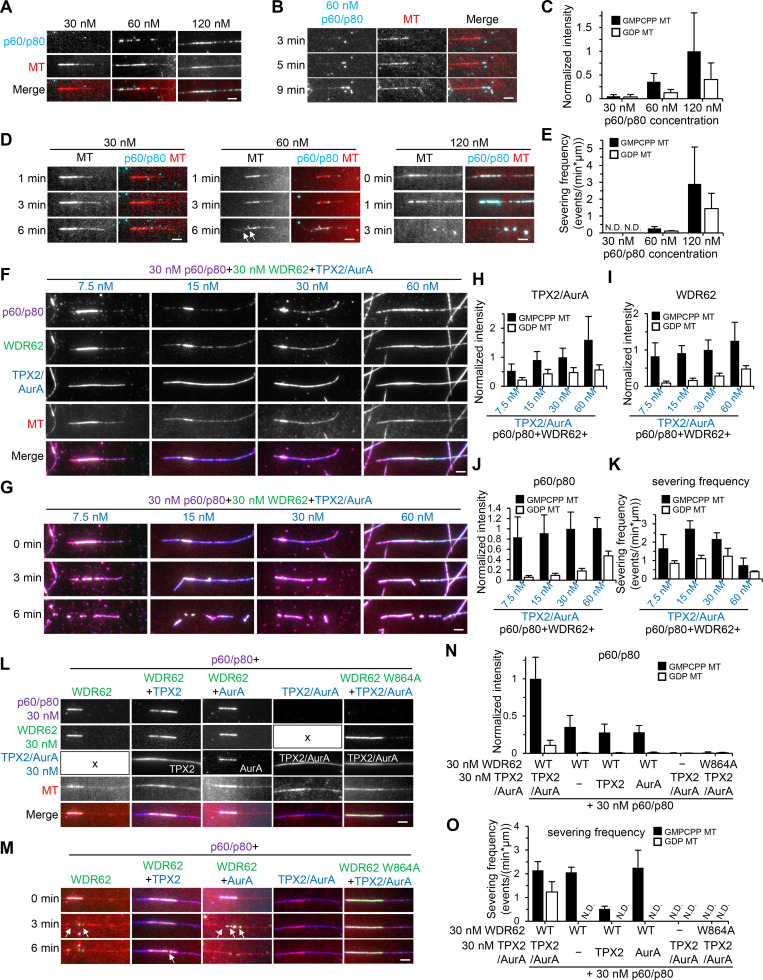
Finally, we set out to investigate the overall activity of the TPX2–Aurora A–WDR62–katanin module in vitro. We first performed a titration in katanin concentration from 30 nM to 120 nM in the dynamic MT assays. We observed that its MT-binding affinity and severing frequency both increased in a dose-dependent manner. Moreover, its affinity for GMPCPP seeds was stronger than that of GDP-MTs; hence, severing events were more frequently observed along seeds (Fig. 9, A and C–E). Consistent with our previous work, the FL katanin also showed MT end binding (Fig. 9 B).
Figure 9.
The TPX2–Aurora A–WDR62–katanin module can potently disassemble GDP-MTs in vitro.(A–C) Images and quantification of 30 nM, 60 nM, and 120 nM p60/SNAP-p80 binding to preassembled dynamic MTs in flow-in experiments. At 60 nM, p60/p80 was also observed to bind and bend the MT end (B). n = 19–22 MTs from two experiments. (D) Time-lapse images showing no severing, a few severing events on GMPCPP seed (arrows), and efficient severing of preassembled dynamic MTs in flow-in experiments with 30 nM, 60 nM, and 120 nM p60/SNAP-p80, respectively. (E) Severing frequency of p60/p80 on GMPCPP seeds and GDP MTs at indicated concentrations. n = 2 experiments; 19–22 MTs were measured. (F) Images showing the recruitment of 30 nM p60/SNAP-p80 to dynamic MTs preassembled in the presence of 30 nM WDR62-GFP and indicated concentration of TPX2/TagBFP–Aurora A (AurA) in flow-in experiments. (H–J) Quantification of intensities of TPX2/Aurora A (H), WDR62 (I), and p60/p80 (J) on GMPCPP seeds and GDP MTs immediately after flow-in as shown in F. Values were normalized to the intensity in the condition with 30 nM TPX2/TagBFP–Aurora A, 30 nM WDR62-GFP, and 30 nM p60/SNAP-p80 on GMPCPP seeds. n = 20–30 MTs from two experiments. (G and K) Images and quantification of MT-severing frequency of 30 nM p60/SNAP-p80 in the presence of 30 nM WDR62-GFP and indicated concentration of TPX2/TagBFP–Aurora A. n = 2 experiments; 20–30 MTs were measured. (L and N) Images and quantification of intensities of p60/p80 on GMPCPP seeds and GDP MTs in the presence of indicated factors. All proteins were used at 30 nM. The "x" in L indicates the absence of TPX2/AurA or WDR62 in the corresponding assays. Values were normalized to the intensity in the condition with 30 nM TPX2/TagBFP–Aurora A, 30 nM WDR62-GFP, and 30 nM p60/SNAP-p80 on GMPCPP seeds. n = 20–30 MTs from two experiments. (M and O) Images and quantification of MT-severing frequency of p60/p80 on GMPCPP seeds and GDP MTs in the presence of indicated factors at the same concentrations as in L. Arrows indicate severing events on GMPCPP seeds. Of note, the severing frequency of p60/p80 on seeds in the presence of WDR62 and TPX2 was lower than that in the presence of WDR62 alone or WDR62 and Aurora A, which reflects the stabilization effect exerted by TPX2. n = 2 experiments; 20–30 MTs were measured. Scale bars, 2 µm. Data represent mean ± SD. N.D., not detected.
Since no severing was observed at 30 nM katanin, to highlight the contribution of TPX2/Aurora A in subsequent assays, both katanin and WDR62 were used at a fixed concentration of 30 nM, while TPX2/Aurora A was used over a range of concentrations from 7.5 nM to 60 nM. Increasing concentrations of TPX2/Aurora A caused the expected increased recruitment of katanin to both seeds and GDP-MTs (Fig. 9, F and H–J). Surprisingly, the severing frequency peaked at moderate concentrations and dropped at high concentrations of TPX2/Aurora A (Fig. 9, G and K; compare severing in the presence of 30 nM and 60 nM TPX2/Aurora A in Video 6). We reasoned that TPX2 can not only indirectly recruit katanin to sever MTs but can also stabilize MTs by itself. Both of these two opposing effects increase with increasing TPX2/Aurora A concentration; only at moderate concentrations could the effect of katanin-mediated severing predominate over the stabilization effect, leading to the peak severing frequency. For controls, we also examined the severing activity of 30 nM katanin in the presence of three other factors either alone, combined in pairs, or altogether, but using W864A mutant instead of WT WDR62; as expected, when WT WDR62 was present, katanin could be recruited to sever seeds; however, in all cases, recruitment of katanin to GDP-MTs was hardly observed, and hence no severing occurred there (Fig. 9, L–O). Collectively, we conclude that TPX2, Aurora A, WDR62, and katanin act together as a highly efficient module for disassembly of GDP-MTs, which underlies the precise regulation of spindle MT dynamics.
Video 6.
TPX2, Aurora A, WDR62, and katanin form an efficient module to sever GDP-MTs in vitro. MTs were first polymerized in the tubulin polymerization reaction mixture (20 µM unlabeled tubulin and 0.5 µM rhodamine-tubulin in MRB80 buffer) supplemented with 30 nM WDR62-GFP and 30 nM (top panels) or 60 nM (bottom panels) TPX2/TagBFP–Aurora A (AurA). After reaction for 10 min on the microscope stage (30°C), the sample in the unsealed flow chamber was imaged for ∼1 min. Subsequently, the second-round reaction mixture, which contained 10 µM tubulin together with 30 nM p60/SNAP-Alexa Fluor 647–p80, 30 nM WDR62-GFP, 30 nM (top panels) or 60 nM (bottom panels) TPX2/TagBFP–Aurora A, and 1 mM ATP was gently flowed into the chamber. Images were collected with a TIRF microscope at 10-s intervals. Video is sped up 60 times (6 frames/s). Time is shown in the format min:s.
Discussion
In this study, we unveil WDR62 as an atypical MAP presumably recognizing extended MT lattice. This unique property makes it unlikely to directly regulate spindle MT dynamics. Rather, WDR62 functions as an adaptor protein between its recruiting factor TPX2/Aurora A and the effector katanin to regulate MT dynamics at spindle poles. Mechanistically, reconstitution assays showed that the TPX2–Aurora A–WDR62–katanin axis can potently promote disassembly of GDP-MTs in vitro.
TPX2 is a well-established positive regulator of MT assembly, as it can stimulate MT nucleation and directly promote MT stability by slowing down depolymerization and reducing the frequency of catastrophe (Schatz et al., 2003; Roostalu et al., 2015; Wieczorek et al., 2015; Reid et al., 2016; Woodruff et al., 2017). Here, we showed that TPX2 promotes katanin-mediated disassembly of GDP-MTs via the TPX2–Aurora A–WDR62–katanin axis, thus uncovering an unexpected role of TPX2 in negatively regulating MT assembly. In contrast to WDR62 and katanin, which almost exclusively localized at the spindle pole during mitosis, both TPX2 and Aurora A were distributed along the whole spindle, being relatively more prominent at the pole region. In the current study, our in vitro assays showed that WDR62 displays older GDP-MT region bias due to TPX2/Aurora A’s recruitment, and this bias was even stronger than TPX2, due to WDR62’s more persistent accumulation over time. This mechanism may partially contribute to the prominent spindle pole localization of WDR62. Collectively, it is tempting to speculate that while TPX2 generally functions as a positive regulator within spindle, the spindle pole–localized pool of TPX2 together with Aurora A can locally recruit WDR62 and katanin to negatively regulate MT assembly, which might contribute to the homeostasis of spindle.
As shown in Fig. 2, we described a series of defects in spindle organization and dynamics in both WDR62 and p80 knockout HeLa cells. This prompted us to consider possible links between these defects. Our data demonstrate that the lack of WDR62 or katanin results in less efficient minus-end depolymerization and, as a consequence, leads to more stable spindle MTs. We further envision that the more stable spindle MTs may affect the activity of minus end–directed motors involved in cross-linking between the centrosome and the spindle pole, such as the dynein complex and/or HSET (Gaglio et al., 1997; Goshima et al., 2005; Chavali et al., 2016), which in turn leads to inward displacement of the centrosome relative to the spindle pole; the “embedded” centrosome within the spindle pole may cause steric hindrance to astral MTs emanating from the centrosome, thus leading to the reduced number and length of astral MTs.
Recently, the Basto group found that at early neurogenic stages, when neural stem cells (NSCs) undergo proliferative symmetric divisions to expand the NSC pool, spindles display more astral MTs and fewer spindle MTs; in contrast, at late neurogenic stages, when NSCs undergo asymmetric neurogenic divisions to give rise to committed neuronal precursors, spindles display fewer astral MTs and higher spindle MT density and stability (Vargas-Hurtado et al., 2019). In our WDR62 and katanin knockout HeLa cells, the number and length of astral MTs were reduced, but the stability of spindle MTs was increased, which is similar to the spindle morphology at late neurogenic stages. We envision that during the transition from early to late neurogenic stages, the loading of WDR62 and katanin to the spindle pole may be down-regulated to cause a switch in spindle morphology; if this was the case, loss of function of WDR62 and katanin at early neurogenic stages may cause premature neuronal differentiation and hence lead to reduction of the NSC pool and small brain size, which may provide a possible explanation for the MCPH etiology associated with WDR62 and katanin mutations.
In the current study, our systematic analysis revealed that MCPH-related mutations within the WD40 domain generally correlate with perturbations in WDR62 interactions with Aurora A and MT. In addition, the MCPH-associated R863H substitution dramatically reduced WDR62 binding to katanin. These findings may provide useful insight for the mechanistic understanding of the MCPH pathogenesis.
Our previous work showed that ASPM acts in complex with katanin to regulate spindle dynamics, and knockout of ASPM could reduce the spindle pole localization of p80 by ∼50% (Jiang et al., 2017). By contrast, in the current study, knockout of WDR62 dramatically reduced the spindle pole localization of p80 by ∼90%, suggesting that WDR62 might be the major recruiting factor of katanin in our system. In agreement with our results, an accompanying study from the Meraldi laboratory also demonstrated that WDR62 can localize katanin at spindle poles to ensure efficient minus-end depolymerization (Guerreiro et al., 2021). Although our studies revealed two pathways (ASPM and WDR62) for recruiting katanin to the spindle pole in HeLa cells, how they work together needs further investigation.
By combining live-cell imaging with in vitro assays, we discovered several lines of evidence supporting that WDR62 recognizes extended MT lattice. This mode of MT binding is unique to the best of our knowledge. MT buckling is a well-documented phenomenon (Waterman-Storer and Salmon, 1997; Janson et al., 2003; Brangwynne et al., 2006; Robison et al., 2016). However, despite great progress toward a better understanding of this subject from biophysical aspects, its cellular function and the underlying mechanisms remain enigmatic. Our identification of WDR62 as a specific marker for curved MTs will provide a useful tool for future functional and mechanistic studies on this subject.
It should be noted, however, that in the WDR62-GFP-Strep knock-in HeLa cells used in this study, WDR62 was dispersed in the cytoplasm and no binding to curved MTs could be observed in interphase, maybe because of the low endogenous expression levels (data not shown). Thus, direct experimental evidence for the contribution of WDR62’s intrinsic affinity for curved MTs is currently lacking in our system. Despite this, since fly WDR62 also shows preference for curved MTs, it suggests that this property is conserved among species and hence may function in other cell types or developmental contexts. Previously, the Black group reported that DCX associates with straight but not curved MTs in the neuronal growth cone and that depletion of DCX family members results in MTs becoming more curvy. They propose that DCX may promote MT straightness to support its function in axonal transport (Jean et al., 2012). In the future, it will be interesting to investigate whether WDR62 could be observed to specifically bind to curved MTs in the growth cone and further promote katanin-mediated severing there, thus also contributing to the directional axonal transport.
Materials and methods
DNA constructs
Katanin p60 (NM_001083961) was cloned into Bio-GFP-C1 vector and pTT5 vector without tag, and katanin p80 (NM_028805) was cloned into Bio-GFP-C1, pTagBFP-C1, Strep-C1, and Strep-SNAP-C1 vectors. Human WDR62 (NM_001083961), Aurora A (NM_001323303.2), and TPX2 (NM_012112) were amplified from HeLa cDNA. Full-length WDR62 and its truncations were cloned into EGFP-Strep-N1, EGFP-N1, mCherry-N1, EGFP-C1, Bio-GFP-C1, and pET-32a (+) vectors. Aurora A was cloned into pTT5-Strep-GFP-C1, pTT5-Strep-TagBFP-C1, and pET-Strep-GST-C1 vectors. TPX2 was cloned into pTT5-Strep-TagBFP-C1 vector and pTT5 vector without tag. Human DCX (NM_001369371.1) was a gift of Jiahuai Han (Xiamen University, Xiamen, China) and cloned into EGFP-N1 and mCherry-N1 vectors. Fly WDR62 was amplified from a Drosophila cDNA clone (ID: LD01189; Drosophila Genomics Resource Center) and cloned into EGFP-C1 vector.
Cell culture and transfection
All cell lines were cultured in DMEM/F12 (1:1; Hyclone) supplemented with 10% FBS and 5 U/ml penicillin and 50 µg/ml streptomycin and kept at 37°C in 5% CO2. FuGENE6 (Promega) was used to transfect plasmids into HeLa, MRC5, or U2OS cells for knockdown, knock-in, immunofluorescence, and live-cell imaging; polyethylenimine (PEI; Polysciences) was used to transfect plasmids into HEK293T cells for protein purification and pull-down experiments.
Antibodies
The following primary antibodies were used: rabbit polyclonal antibodies against GFP (Proteintech; 50430–2-AP), katanin p80 (Proteintech; 14969–1-AP), WDR62 (Thermo Fisher Scientific; A301-560A), TPX2 (Proteintech; 11741–1-AP), NuMA (Thermo Fisher Scientific; PA3-16829), KIF2A (Proteintech; 13105–1-AP), KIF2C (ABclonal; A5449), and α-tubulin (Proteintech; 11224–1-AP); a rabbit monoclonal antibody against Aurora A (CST; 4718); mouse monoclonal antibodies against β-tubulin (Sigma-Aldrich; T5201), γ-tubulin (Thermo Fisher Scientific; MA1-19421), and Strep II-Tag (ABclonal; AE066); and a rat monoclonal antibody against α-tubulin YL1/2 (Thermo Fisher Scientific; MA1-80017).
Generation of stable cell lines
To generate WDR62 knockout HeLa cell line, cells grown on six-well plates were transfected with 2 µg PX459 bearing single guide RNA (5′-GTGTCACTCGAGAAGGTGCT-3′) and selected with 8 µg/ml puromycin for 2 d. 4–5 d after removal of puromycin, cells were diluted in 96-well plates to isolate single-cell colonies. Positive clones were screened by immunofluorescence and Western blotting. Katanin p80 knockout HeLa cell line (gRNA: 5′-CGTCCGCCTCAACACCCCCG-3′) was generated in a similar way and characterized previously (Jiang et al., 2017).
To tag WDR62 locus with GFP at the C-terminus, 2 µg donor construct harboring a GFP-Strep-T2A-blas cassette was cotransfected with 1 µg PX459 bearing single guide RNA (5′-GCCGTGCGGAGGAAGGCACG-3′) into HeLa cells grown on six-well plates. Cells were first selected with 8 µg/ml puromycin for 2 d and then selected with 5 µg/ml blasticidin for 1 wk, followed by serial dilution in 96-well plates. Positive clones were screened by immunofluorescence and Western blotting. The KIF2C-GFP-Strep knock-in HeLa cell line (gRNA: 5′-TTTTATTTGCAGTCGTCACT-3′) was generated in a similar way. For previously described U2OS-PA-GFP-tubulin-EB3-TagRFP cell line (Jiang et al., 2017), a tandem cytomegalovirus cassette expressing PA-GFP-tubulin and EB3-TagRFP was stably integrated into the AAVS1 locus (gRNA: 5′-GTCACCAATCCTGTCCCTAG-3′).
For genotyping of WDR62 knockout HeLa cell line, WDR62 knockout genotyping-forward primer (5′-TTCTCTCTTGCACCCATCATTACTTG-3′) and WDR62 knockout targeting sequence-reverse primer (5′-TCCCAGAATCGCTGTCACCT-3′) were used (Fig. S1 E and Table S1). For genotyping of WDR62- or KIF2C-GFP-Strep knock-in HeLa cell lines, 5′ homology arm-forward primers and 3′ homology arm-reverse primers (WDR62: 5′-GCATTGGAGCAGAGACCTGT-3′ and 5′-CCCTGTTGCAGATGGGGACAC-3′; KIF2C: 5′-ATGGGGATAAGCTTGGGATAGGA-3′ and 5′-GTTCCCAGGCCTAGTAAGCA-3′) were used (Fig. S1, B and P; and Table S1).
Generation of Auxin (IAA)-inducible degradation cell lines
To establish cell lines for Auxin-inducible degradation of TPX2 or Aurora A, we took a two-step approach. We first generated the Tet-3xHA-OsTIR1-puro parental HeLa cell line. pMK243 plasmid (Addgene; #72835) was modified to generate the donor construct for inducible expression of 3xHA-OsTIR1 at AAVS1 locus. 2 µg donor construct and 1 µg PX330 bearing single guide RNA (5′-GGGGCCACTAGGGACAGGAT-3′) were cotransfected into HeLa cells grown on six-well plates. 2 d after transfection, the cells were selected with 1 µg/ml puromycin for 2 d followed by serial dilution after withdrawing drug selection. The positive clones were selected based on the expression level of 3xHA-OsTIR1 in the presence of doxycycline as assessed by immunofluorescence staining and Western blotting. Subsequently, the TPX2 or Aurora A locus was further tagged with the mini AID (mAID)-mClover-hygro cassette at the C-terminus in the parental cell line via CRISPR/Cas9 (gRNA for TPX2: 5′-CTCACAGCTGAGTTTAGCAG-3′; gRNA for Aurora A: 5′-CAGTCTTAGGAATCGTGCAG-3′). For this, 2 µg donor construct and 1 µg PX330 bearing single guide RNA were cotransfected into the parental cell line grown on a six-well plate. 3 d after transfection, the cells were selected with 200 µg/ml hygromycin B for 1 wk followed by serial dilution after withdrawing drug selection. Immunofluorescence staining was performed to select positive clones, and further Western blotting was performed to select the homozygous knock-in clones. The 5′ homology-arm forward primers and 3′ homology-arm reverse primers (TPX2: 5′-TGCCTTCTTTAGCCACCCTTC-3′ and 5′-GTGAGGCTCCCCATGAATCC-3′; Aurora A: 5′-TGTCATTGCGACCAGGCTTT-3′ and 5′-TAACAGCTCTGAGACACATGGC-3′) were used for genotyping (Fig. S4 C and Table S1).
The established Aurora A- or TPX2-mAID-mClover knock-in HeLa cell lines were first treated with 2 µg/ml doxycycline for 12 h to induce the expression of OsTIR1. Subsequently, cells were treated with 2 µg/ml doxycycline and 500 µM IAA for 6 h (Fig. 7, A, B, D, and E; and Fig. S4, K and L) or 12 h (Fig. S4, D–J) to induce the degradation of Aurora A– or TPX2-mAID-mClover. For Western blotting analysis in Fig. S4 D, 10 µM S-Trityl-L-cysteine (STLC) was added together with 2 µg/ml doxycycline and 500 µM IAA for 12 h before harvesting to synchronize cells in mitosis.
Protein expression and purification from HEK293T cells
For protein overexpression in HEK293T cells, EGFP-Strep-N1 vector was used for WDR62 FL and WD40 domain–containing fragments and pTT5 vector for other proteins.
Generally, HEK293T cells grown on 15-cm dishes were transfected with 20–30 µg DNA per dish using PEI. Cells were treated with 200 ng/ml nocodazole overnight before harvesting. 36 h after transfection, the medium was removed from the dishes, and cells were collected with cold PBS (4°C, 10 ml for each 15-cm dish) into 15-ml falcon tubes quickly. Cells were centrifuged at 1,000× rpm, 4°C for 10 min to remove the supernatant, and the pellets were lysed in 900 µl lysis buffer (50 mM Hepes, 300 mM NaCl, and 0.5% Triton X-100, pH 7.4) containing protease inhibitors (Roche) for 10 min on ice. Cell lysate was centrifuged at 14,000× rpm, 4°C for 20 min, and the supernatant was incubated with 60∼100 µl StrepTactin beads (GE Healthcare) at 4°C for 45 min. After removal of the supernatant by centrifuging at 3,000× rpm, 4°C for 1 min, beads were washed with 1 ml lysis buffer four times and 1 ml wash buffer A (50 mM Hepes, 150 mM NaCl, and 0.01% Triton X, pH 7.4) two times. Finally, proteins were eluted with 60∼100 µl elution buffer (50 mM Hepes, 150 mM NaCl, 0.01% Triton X-100, and 2.5 mM desthiobiotin, pH 7.4), snap frozen, and stored at −80°C.
To copurify the TPX2/Aurora A complex, nontagged pTT5-TPX2 was cotransfected with pTT5-Strep-TagBFP–Aurora A at a ratio of 1 to 1. The complex was purified as described above.
To copurify the katanin complex, Strep-SNAP-katanin p80 was cotransfected with nontagged pTT5-katanin p60 at a ratio of 1 to 1.5. Cells were collected and lysed as described above. After incubation with the supernatant, beads were washed with lysis buffer four times and wash buffer A two times as described above. Subsequently, to label SNAP-tag proteins, beads were incubated with 100 µl wash buffer A containing 10 µM SNAP-Surface Alexa Fluor 647 (NEB) at 4°C for 1 h in darkness. After extensive washing with lysis buffer four times and wash buffer B (50 mM Hepes, 300 mM NaCl, and 0.01% Triton X, pH 7.4) two times, proteins were eluted with 60 µl elution buffer containing 300 mM instead of 150 mM NaCl.
The yield of Strep-TagBFP-TPX2, Strep-TagBFP–Aurora A, Strep-GFP–Aurora A, and Strep-TagBFP–Aurora A/TPX2 was ∼35–75 µg per 15-cm dish. The yield of WDR62 FL WT-GFP-Strep with or without JNK inhibitor treatment, WDR62 FL W864A-GFP-Strep, WDR62 WD-GFP-Strep, Strep-GFP-WDR62 ΔWD, WDR62 FL T1053A-GFP-Strep, and p60/Strep-SNAP-p80 was ∼2.5–5 µg per 15-cm dish. Of note, in the case of WDR62 FL WT-GFP-Strep with JNK inhibitor treatment, cells were treated with 10 µM JNK inhibitor VIII (Cayman; 15946) together with 200 ng/ml nocodazole overnight before harvesting; the lysis buffer was also supplemented with 10 µM JNK inhibitor VIII.
All purified proteins described above were analyzed by SDS-PAGE (Fig. S5 G).
Protein expression and purification from Escherichia coli
For protein overexpression in E. coli, BL21 (DE3) strain was used. Aurora A was cloned into a modified pET28a vector (Novagen) containing a GST tag. GST control was expressed from the pGEX 6p-1 vector (GE Healthcare). GST–Aurora A and GST control were purified with Glutathione Sepharose 4 fast flow beads (GE Healthcare). Beads in lysis buffer containing 50% (vol/vol) glycerol were snap frozen and stored at −80°C for subsequent GST pull-down assays. WDR62 M1 WT and W864 mutant were cloned into pET32a vector with a trxA-6xHis tag. trxA-6xHis-WDR62 M1 proteins were purified with Ni-NTA agarose (Qiagen). Proteins were eluted with 500 µl elution buffer containing 250 mM imidazole, snap frozen, and stored at −80°C. Purification conditions and procedures were performed according to the manual provided by the vendors.
Pull-down assays
For StrepTactin pull-down assays (Fig. 3 D, Fig. 7 J, and Fig. S5 C), HEK293T cells seeded on six-well plates were cotransfected with 1 µg Strep- or Strep-GFP–tagged bait construct and 1 µg GFP-tagged prey construct (2 µg total DNA) using PEI. 36 h after transfection, cells were collected and lysed in 100 µl lysis buffer for 5 min on ice. Cell lysate was centrifuged at 14,000× rpm, 4°C for 20 min, and the supernatant was incubated with 10 µl StrepTactin beads at 4°C for 45 min. Subsequently, beads were washed with lysis buffer four times and then subjected to Western blotting.
For the StrepTactin pull-down assay in Fig. 3 G, HEK293T cells seeded on six-well plates were transfected with 2 µg Strep-GFP or Strep-p80 WD. Strep-tagged proteins immobilized on StrepTactin beads were used as bait. trxA-6xHis-WDR62 M1 proteins purified from E. coli as described above were used as prey. Approximately 3 µg prey proteins diluted in 100 µl lysis buffer was incubated with bait-bound beads at 4°C for 45 min. Subsequently, beads were washed with lysis buffer four times and then subjected to SDS-PAGE.
For streptavidin pull-down assays (Fig. 3, B and F; and Fig. 5, D and E), HEK293T cells seeded on six-well plates were cotransfected with 1 µg Bio-GFP–tagged bait construct, 1 µg GFP-tagged prey construct, and 1 µg BirA construct (3 µg total DNA) using PEI. 36 h after transfection, cells were collected and lysed in 100 µl lysis buffer for 5 min on ice. Cell lysate was centrifuged at 14,000× rpm, 4°C for 20 min to collect the supernatant. Streptavidin-coated magnetic beads (Invitrogen; Dynabeads M-280; 15 µl for each sample) were preblocked with 500 µl block buffer (0.1% Triton X-100 and 1 mg/ml chicken egg white in PBS) for 30 min at room temperature. The supernatant was incubated with Dynabeads at 4°C for 45 min. Subsequently, beads were washed with lysis buffer four times and then subjected to Western blotting. Of note, in the case of JNK inhibitor treatment (Fig. 5 E), cells were treated with 10 µM JNK inhibitor VIII overnight before harvesting; the lysis buffer was also supplemented with 10 µM JNK inhibitor VIII.
For GST pull-down assays (Fig. 7 H), HEK293T cells seeded on 10-cm dishes were transfected with 10 µg GFP-tagged prey construct using PEI. 36 h after transfection, cells were collected and lysed in 300 µl lysis buffer for 10 min on ice. Cell lysate was centrifuged at 14,000× rpm, 4°C for 20 min to collect the supernatant. GST control and GST–Aurora A (bait) were purified from E. coli and immobilized on beads as described above. The beads were washed with lysis buffer two times and then incubated with the supernatant at 4°C for 45 min. Subsequently, beads were washed with lysis buffer four times and then subjected to Western blotting.
In vitro MT assays
Double-cycled GMPCPP MT seeds were made as described previously (Mohan et al., 2013). Briefly, 8.25 µl tubulin reaction mixture in MRB80 buffer (80 mM Pipes, 1 mM EGTA, and 4 mM MgCl2, pH 6.8), which contained 14 µM unlabeled porcine brain tubulin (Cytoskeleton), 3.6 µM biotin-tubulin (Cytoskeleton), 2.4 µM rhodamine-tubulin (Cytoskeleton), and 1 mM GMPCPP (Jena Biosciences), was incubated at 37°C for 30 min. MTs were then pelleted by centrifugation in an Airfuge (Beckman) at ∼28 psi for 5 min. After the supernatant was carefully removed, the pellet was resuspended in 6 µl MRB80 buffer and depolymerized on ice for 20 min. Subsequently, a second round of polymerization was performed at 37°C in the presence of freshly supplemented 1 mM GMPCPP. The MT seeds were then pelleted as described above and resuspended in 50 µl MRB80 buffer containing 10% glycerol, snap frozen, and stored at −80°C.
Flow chambers for assays were made of plasma-cleaned glass coverslips and microscope slides. The flow chambers were sequentially incubated with 0.2 mg/ml Poly (L-lysine)-poly (ethylene glycol) (PLL-PEG)-biotin (Susos AG) and 1 mg/ml neutravidin (Invitrogen) in MRB80 buffer. GMPCPP seeds were then attached to coverslip via biotin-neutravidin links, followed by blocking with 1 mg/ml κ-casein. The reaction mixture, which consisted of purified protein and MRB80 buffer containing 20 µM porcine brain tubulin, 0.5 µM rhodamine-tubulin, 1 mM GTP, 0.2 mg/ml κ-casein, 0.1% methylcellulose, and oxygen scavenger mix (50 mM glucose, 400 µg/ml glucose oxidase, 200 µg/ml catalase, and 4 mM DTT), was added to the chamber after centrifugation in an Airfuge for 5 min at ∼28 psi. The flow chamber was sealed with vacuum grease and imaged immediately at 30°C using a TIRF microscope. The imaging interval was 3 s unless stated otherwise.
For all severing assays (Fig. 6, E and G; and Fig. 9), additional 1 mM ATP was added into the reaction mixture. For severing assays of dynamic MTs, GMPCPP seeds were first elongated in the presence of the first-round reaction mixture, which contained 20 µM tubulin (Fig. 9, A, B, and D) or 20 µM tubulin together with the indicated proteins except for katanin (Fig. 9, F, G, L, and M). After reaction for 10 min on the microscope stage (30°C), the sample in the unsealed flow chamber was imaged for ∼1 min. Subsequently, the second-round reaction mixture, which contained 10 µM tubulin together with all indicated proteins including katanin, was gently flowed into the chamber. The imaging interval was 10 s.
For assays with nondynamic MTs, tubulin was excluded from the reaction mixture. The imaging interval was 10 s.
For assays aiming to generate curved MTs (Fig. 4 H), GMPCPP seeds were first elongated in the presence of 20 µM tubulin for 5 min. Subsequently, the reaction mixture containing 30 nM purified WDR62 and 10 µM tubulin was quickly flowed into the chamber with preassembled dynamic MTs. MT buckling appeared frequently because of the mechanical strain cause by the solution exchange.
TIRF microscopy
TIRF microscopy was performed on Nikon Eclipse Ti2-E with the perfect focus with the Nikon CFI Apo TIRF 100× 1.49 NA oil objective, Prime 95B camera (Photometrics), SOLE laser engine (four lasers: 405 nm, 488 nm, 561 nm, and 638 nm; Omicron) and controlled by NIS-Elements software (Nikon). Images were magnified with a 1.5× intermediate lens on Ti2-E before projected onto the camera. The resulting pixel size is 73.3 nm/pixel. Stage top incubator INUBG2E-ZILCS (Tokai Hit) was used to keep cells at 37°C or in vitro samples at 30°C. Imaging medium (DMEM/F12 supplemented with 10% FBS and 5 U/ml penicillin and 50 µg/ml streptomycin) was prewarmed in a water bath at 37°C.
Optosplit III beamsplitter (Cairn Research Ltd.) was used for simultaneous imaging of green and red fluorescence. Stream acquisition was used for simultaneous imaging of green and red fluorescence in vivo. Sequential acquisition was used for three- or four-color imaging experiments.
Photoactivation
The microscope setup for photoactivation experiments was the same as that for TIRF microscopy except that the epi-fluorescence illuminator was used (Intensilight; Nikon) and the 1.5× intermediate lens was omitted. Photoactivation of PA-GFP-tubulin at designated regions in the spindle was achieved using Nikon Galvo Miniscanner with a 405-nm laser.
Immunofluorescence
Cells for immunofluorescence were seeded on coverslips in 24-well plates and transfected with 0.5–0.75 µg plasmid using FuGENE6. 18 h after transfection, cells were fixed with different methods. For staining of astral MTs in Fig. 2 A and Fig. S2 B and for staining of cold-resistant spindle MTs in Fig. 2 L, cells were fixed in 4% formaldehyde and 0.1% glutaraldehyde in 37°C prewarmed MRB80 buffer for 10 min at room temperature. For staining of MTs in Fig. S4, E and F and Fig. S5, A and F, cells were sequentially fixed in −20°C methanol for 5 min and 4% formaldehyde in PBS for 5 min at room temperature. For staining of other proteins, cells were fixed with −20°C methanol for 10 min. Cell membranes were permeabilized with 0.15% Triton-X 100 in PBS, and subsequent blocking and labeling steps were performed in PBS supplemented with 2% BSA and 0.05% Tween-20. The permeabilizing, blocking, and labeling steps were performed at room temperature. Finally, coverslips were rinsed with 70% and 100% ethanol, air-dried, and mounted on glass slides with Vectashield mounting medium (Vector Laboratories). Slides were stored at −20°C.
Images were captured using Nikon Ni-U with 60× 1.40 NA oil objective equipped with DS-Qi2 camera (Nikon) for single-slice acquisition or Nikon Eclipse Ti2-E with 100× 1.45 NA oil objective equipped with Prime 95B camera for 3D acquisition with 0.11 µm z steps.
Cold treatment
Cells were seeded on coverslips in 24-well plate for 18 h. The plate was cooled in an ice-water bath for 5 min, followed by immunofluorescence staining of cold-resistant spindle MTs (Fig. 2 L), which was described above.
Image analysis and processing
For measurement of the intensity of indicated proteins at the spindle pole (Fig.1 E; Fig. 3 I; Fig. 7, C and F; Fig. S1, H and M; and Fig. S4, L, O, and P), a trapezoid region of interest (ROI) was selected to cover the spindle pole structure. For measurement of the intensity of KIF2C and γ-tubulin at the centrosome (Fig. S1, I and N), a circular ROI was selected to cover the centrosome structure.
For measurement of “inside” and “outside” intensities of NuMA (spindle pole marker) and γ-tubulin (centrosome marker) in Fig. 2 H and Fig. S2 F, images were rotated to orient the spindles with a horizontal centrosome-to-centrosome axis. Two semicircular ROIs generated by the ImageJ macro HalfCircleSelections formed a circle to cover the whole spindle pole structure. The center of the circle was placed at the visually determined center of the γ-tubulin spot. The chromosome- and cortex-directed ROIs were referred to as inside and outside, respectively. The outside versus total intensity of γ-tubulin was ∼0.5 on average, indicating that the visually determined center of the γ-tubulin spot is reliable. The same ROIs were used to measure the inside and outside intensities of NuMA.
For measurement of the integrated intensity of the cold-resistant spindle MTs (Fig. 2 M), a polygon ROI was selected to cover the spindle. The intensity of each spindle was averaged from the integrated intensity of the four middle Z-stacks. The number and length of astral MTs were measured manually in ImageJ using the maximum projection image of Z-stacks (Fig. 2, B and C; and Fig. S2, C and D).
The center of the γ-tubulin spot and the distal edge of the NuMA spot were determined visually in ImageJ. The tcentrosome-to-centrosome distance was defined as the center-to-center distance between two γ-tubulin spots, and the pole-to-pole distance was defined as the distal edge–to–distal edge distance between two NuMA spots (Fig. 2 F). The distance between the centrosome and spindle pole was defined as the distance between the center of the γ-tubulin spot and the distal edge of the NuMA spot (Fig. 2 G and Fig. S2 G).
For measurement of the intensity of indicated proteins in the in vitro assays, a 5-pixel-wide linear ROI was drawn along the MT at the start of imaging (Fig. 4 F; Fig. 5, B and G; and Fig. 6 F), or immediately after flow-in (Fig. 4 I; and Fig. 9 C, H–J, and N), or after 5 min of GDP-MT growth from seed, when the signals of indicated proteins reached a plateau (Fig. 8, F, I, and L). For plots of the intensity of indicated proteins against time (Fig. 8, C, G, and K), a 2-µm-long segment of GDP-MT, as illustrated in Fig. 8 A (between two dashed lines), was tracked for 3 min or 6 min with a time interval of 10 s.
For fluorescence intensity profiles (Fig. 2 D and Fig. 4 D), the line scan was performed using the profile function in ImageJ. To analyze the relative distribution of γ-tubulin and NuMA, a 1-pixel-wide line was drawn along the centrosome-to-centrosome axis, in which the intensities of γ-tubulin and NuMA were measured. The γ-tubulin intensity was normalized to its maximum value for each centrosome, and similarly, the NuMA intensity was normalized to its maximum value for each spindle pole (Fig. 2 D). To analyze the relative distribution of WDR62 and DCX, a 5-pixel-wide line was drawn along the MT (Fig. 4 D).
Surface function in Imaris was used to rebuild the 3D reconstruction. The length of the grid in the image is 3.65 µm (Fig. 2 E).
Severing frequency in vitro was determined as the observed number of MT-severing events divided by the product of the MT length and the observation time. For severing of GMPCPP-stabilized MTs (Fig. 6 H; and Fig. 9, E, K, and O, GMPCPP MT), the MT length was the initial length measured at 0 min (the start of imaging). For severing of dynamic GDP-MTs at high frequency in flow-in assays (Fig. 9, E and K, 120 nM), the MT length was the initial length immediately measured after flow-in. For severing of dynamic GDP-MTs at low frequency in flow-in assays (Fig. 9, E and O, 30 nM and 60 nM), in which the MTs showed overall growth, the MT length was the average of lengths measured immediately after flow-in and at the end of imaging. The imaging windows for nondynamic MT assays and dynamic MT assays were 10 min and 8 min, respectively.
The MT breakage frequency in cells was determined as the number of curved MT breakage events divided by the total number of WDR62-decorated curved MTs (Fig. 6 C). The lag time between appearance of bending and breakage was counted from the time of WDR62 loading onto curved MTs to the time of breakage (Fig. 6 D).
The minus-end depolymerization rate (Fig. 2 K) was measured using an ImageJ macro. Briefly, the position of the spindle pole was determined as the center of a circular ROI that enclosed the tubulin signal at the centrosome. A polygon ROI was selected to cover the PA-GFP–α-tubulin stripe. Each pixel within the polygon ROI was projected onto the pole-to-pole axis, and the average distance between the projected pixels and the pole was measured. Measurements were conducted at seven time points with a 0.5∼1–min interval. Linear regression was then performed to calculate the minus-end depolymerization rate.
All intensity measurements in this study were background-subtracted. “Kappa,” an ImageJ plugin, was used to quantify the MT curvature, |κ|, and protein intensity on both curved and straight MTs (Bechstedt et al., 2014). Kymograph was generated with an ImageJ plugin, KymoResliceWide. 3D image stacks were deconvolved using Huygens Essential (SVI). Images were prepared for publication using ImageJ and Adobe Photoshop.
Identification of WDR62-interacting proteins by mass spectrometry
To identify WDR62-interacting proteins, HEK293T cells were cotransfected with WDR62-GFP-Bio and BirA and treated with 200 ng/ml nocodazole for 16 h before harvesting. WDR62-GFP-Bio and its associated proteins were pulled down by streptavidin beads and run on SDS-PAGE gel for 1 cm. After the gel was stained with Coomassie dye G-250, the lane was cut and subjected to mass spectrometry using UHR-QqTOF mass spectrometer (Bruker) and PEAKS 8.5 software (BSI).
Statistical analysis
Statistical analysis was performed with Excel (Microsoft). P values were determined by unpaired two-tailed t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 characterizes the WDR62 knock-in and knockout cell lines. Fig. S2 shows the overview of WDR62 deletion mutants used in this study and the rescue of spindle defects by WT WDR62-GFP but not its katanin binding–deficient mutants. Fig. S3 shows that paclitaxel alters the MT lattice preference of WDR62 and katanin disassembles WDR62-decorated curved MT lattices. Fig. S4 characterizes the TPX2- or Aurora A–mAID-mClover knock-in cell lines. Fig. S5 shows that MCPH-associated mutations within the WD40 domain of WDR62 disrupt its interactions with both MTs and Aurora A. Video 1 shows the reduced rate of MT minus-end depolymerization at the spindle pole in katanin p80 and WDR62 knockout cells. Video 2 and Video 3 show that WDR62-GFP and WDR62 WD-GFP preferentially accumulate at curved segments of dynamic MTs in cells. Video 4 shows that WDR62 WD-mCherry and DCX-GFP exhibit opposite responses to MT buckling and straightening. Video 5 shows that WT WDR62 N2-GFP recruits p60/TagBFP-p80 to sever curved MTs in cells. Video 6 shows that TPX2, Aurora A, WDR62, and katanin form an efficient module to sever GDP-MTs in vitro. Table S1 lists the CRISPR-Cas9 guide RNA targeting sequences and PCR primers for generating homology recombination donor constructs and genotyping.
Supplementary Material
presents CRISPR-Cas9 gRNA targeting sequences and PCR primers for generating homology recombination donor constructs and genotyping.
Acknowledgments
We thank Patrick Meraldi (University of Geneva, Geneva, Switzerland) for sharing results prior to publication and discussions.
K. Jiang was supported by grants from National Natural Science Foundation of China (32070705 and 31871356), Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (TFJC2018005), and Fundamental Research Funds for the Central Universities.
The authors declare no competing financial interests.
Author contributions: Conceptualization, K. Jiang, J. Huang, and Z. Liang; Methodology, K. Jiang, S. Hua, J. Huang, and Z. Liang; Resources, C. Guan; Investigation, J. Huang and Z. Liang; Visualization, J. Huang, Z. Liang, S. Hua, and K. Jiang; Writing - Original Draft, K. Jiang, S. Hua, J. Huang, and Z. Liang; Writing - Review and Editing, K. Jiang and S. Hua; Funding Acquisition, K. Jiang; and Supervision, K. Jiang.
References
- Alushin, G.M., Lander G.C., Kellogg E.H., Zhang R., Baker D., and Nogales E.. 2014. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 157:1117–1129. 10.1016/j.cell.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstedt, S., Lu K., and Brouhard G.J.. 2014. Doublecortin recognizes the longitudinal curvature of the microtubule end and lattice. Curr. Biol. 24:2366–2375. 10.1016/j.cub.2014.08.039 [DOI] [PubMed] [Google Scholar]
- Bilgüvar, K., Oztürk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A.O., Gökben S., et al. 2010. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 467:207–210. 10.1038/nature09327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A.W., and Hyman A.A.. 2008. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182:289–300. 10.1083/jcb.200802005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch, M.A., Yeap Y.Y., Qu Z., Ngoei K.R., Yip Y.Y., Zhao T.T., Heng J.I., and Ng D.C.. 2012. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J. Cell Sci. 125:5096–5109. 10.1242/jcs.107326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne, C.P., MacKintosh F.C., Kumar S., Geisse N.A., Talbot J., Mahadevan L., Parker K.K., Ingber D.E., and Weitz D.A.. 2006. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J. Cell Biol. 173:733–741. 10.1083/jcb.200601060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali, P.L., Chandrasekaran G., Barr A.R., Tátrai P., Taylor C., Papachristou E.K., Woods C.G., Chavali S., and Gergely F.. 2016. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat. Commun. 7:11005. 10.1038/ncomms11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.F., Zhang Y., Wilde J., Hansen K.C., Lai F., and Niswander L.. 2014. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5:3885. 10.1038/ncomms4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Katsenelson, K., Wasserman T., Khateb S., Whitmarsh A.J., and Aronheim A.. 2011. Docking interactions of the JNK scaffold protein WDR62. Biochem. J. 439:381–392. 10.1042/BJ20110284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Katsenelson, K., Wasserman T., Darlyuk-Saadon I., Rabner A., Glaser F., and Aronheim A.. 2013. Identification and analysis of a novel dimerization domain shared by various members of c-Jun N-terminal kinase (JNK) scaffold proteins. J. Biol. Chem. 288:7294–7304. 10.1074/jbc.M112.422055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert, T., Le D.T., Link S., Friedmann L., and Woehlke G.. 2012. Spastin’s microtubule-binding properties and comparison to katanin. PLoS One. 7:e50161. 10.1371/journal.pone.0050161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger, A., van Haren J., Ribeiro S.A., and Wittmann T.. 2016. Doublecortin Is Excluded from Growing Microtubule Ends and Recognizes the GDP-Microtubule Lattice. Curr. Biol. 26:1549–1555. 10.1016/j.cub.2016.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio, T., Dionne M.A., and Compton D.A.. 1997. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 138:1055–1066. 10.1083/jcb.138.5.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., Upton K., and Compton D.A.. 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15:1827–1832. 10.1016/j.cub.2005.08.065 [DOI] [PubMed] [Google Scholar]
- Garrett, S., Auer K., Compton D.A., and Kapoor T.M.. 2002. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 12:2055–2059. 10.1016/S0960-9822(02)01277-0 [DOI] [PubMed] [Google Scholar]
- Goshima, G., Nédélec F., and Vale R.D.. 2005. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171:229–240. 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, O.J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Silljé H., Karsenti E., Mattaj I.W., and Vernos I.. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4:871–879. 10.1038/ncb870 [DOI] [PubMed] [Google Scholar]
- Guerreiro, A., De Sousa F., Liaudet N., Ivanova D., Eskat A., and Meraldi P.. 2021. WDR62 localizes katanin at spindle poles to ensure synchronous chromosome segregation. J. Cell Biol. 10.1083/jcb.202007171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, D.J., and Jasper H.. 2019. Control of Intestinal Cell Fate by Dynamic Mitotic Spindle Repositioning Influences Epithelial Homeostasis and Longevity. Cell Rep. 28:2807–2823.e5. 10.1016/j.celrep.2019.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, M.E., de Dood M.E., and Dogterom M.. 2003. Dynamic instability of microtubules is regulated by force. J. Cell Biol. 161:1029–1034. 10.1083/jcb.200301147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, D., Kodani A., Gonzalez D.M., Mancias J.D., Mochida G.H., Vagnoni C., Johnson J., Krogan N., Harper J.W., Reiter J.F., et al. 2016. Microcephaly Proteins Wdr62 and Aspm Define a Mother Centriole Complex Regulating Centriole Biogenesis, Apical Complex, and Cell Fate. Neuron. 92:813–828. 10.1016/j.neuron.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean, D.C., Baas P.W., and Black M.M.. 2012. A novel role for doublecortin and doublecortin-like kinase in regulating growth cone microtubules. Hum. Mol. Genet. 21:5511–5527. 10.1093/hmg/dds395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., Heck A.J.R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19:480–492. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M.2017. Metaphase Spindle Assembly. Biology (Basel). 6:8. 10.3390/biology6010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani, A., Yu T.W., Johnson J.R., Jayaraman D., Johnson T.L., Al-Gazali L., Sztriha L., Partlow J.N., Kim H., Krup A.L., et al. 2015. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. elife. 4:e07519. https://doi.org/ 10.7554/eLife.07519 [DOI] [PMC free article] [PubMed]
- Kufer, T.A., Silljé H.H., Körner R., Gruss O.J., Meraldi P., and Nigg E.A.. 2002. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158:617–623. 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, N.R., Yeap Y.Y., Zhao T.T., Yip Y.Y., Wong S.C., Xu D., Ang C.S., Williamson N.A., Xu Z., Bogoyevitch M.A., and Ng D.C.. 2015. Opposing roles for JNK and Aurora A in regulating the association of WDR62 with spindle microtubules. J. Cell Sci. 128:527–540. 10.1242/jcs.157537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, N.R., Yeap Y.Y., Ang C.S., Williamson N.A., Bogoyevitch M.A., Quinn L.M., and Ng D.C.. 2016. Aurora A phosphorylation of WD40-repeat protein 62 in mitotic spindle regulation. Cell Cycle. 15:413–424. 10.1080/15384101.2015.1127472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., and Roll-Mecak A.. 2018. Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 217:4057–4069. 10.1083/jcb.201612104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Akutsu S.N., Fukumitsu A., Morino H., Masatsuna Y., Hosoba K., Kawakami H., Yamamoto T., Shimizu K., Ohashi H., and Matsuura S.. 2017. PLK1-mediated phosphorylation of WDR62/MCPH2 ensures proper mitotic spindle orientation. Hum. Mol. Genet. 26:4429–4440. 10.1093/hmg/ddx330 [DOI] [PubMed] [Google Scholar]
- Mohan, R., Katrukha E.A., Doodhi H., Smal I., Meijering E., Kapitein L.C., Steinmetz M.O., and Akhmanova A.. 2013. End-binding proteins sensitize microtubules to the action of microtubule-targeting agents. Proc. Natl. Acad. Sci. USA. 110:8900–8905. 10.1073/pnas.1300395110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, A.K., Khurshid M., Désir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A., et al. 2010. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42:1010–1014. 10.1038/ng.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Novorol, C., Burkhardt J., Wood K.J., Iqbal A., Roque C., Coutts N., Almeida A.D., He J., Wilkinson C.J., and Harris W.A.. 2013. Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol. 3:130065. 10.1098/rsob.130065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry, S.2016. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 85:659–683. 10.1146/annurev-biochem-060815-014528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton, C.J., Schot R., Seufert K., Lequin M.H., Accogli A., Annunzio G.D., Villard L., Philip N., de Coo R., Catsman-Berrevoets C., et al. 2014. Severe presentation of WDR62 mutation: is there a role for modifying genetic factors? Am. J. Med. Genet. A. 164:2161–2171. 10.1002/ajmg.a.36611 [DOI] [PubMed] [Google Scholar]
- Qin, Y., Zhou Y., Shen Z., Xu B., Chen M., Li Y., Chen M., Behrens A., Zhou J., Qi X., et al. 2019. WDR62 is involved in spindle assembly by interacting with CEP170 in spermatogenesis. Development. 146:dev174128. 10.1242/dev.174128 [DOI] [PubMed] [Google Scholar]
- Ramdas Nair, A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B., and Cabernard C.. 2016. The Microcephaly-Associated Protein Wdr62/CG7337 Is Required to Maintain Centrosome Asymmetry in Drosophila Neuroblasts. Cell Rep. 14:1100–1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Reid, T.A., Schuster B.M., Mann B.J., Balchand S.K., Plooster M., McClellan M., Coombes C.E., Wadsworth P., and Gardner M.K.. 2016. Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J. Cell Sci. 129:1319–1328. 10.1242/jcs.178806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison, P., Caporizzo M.A., Ahmadzadeh H., Bogush A.I., Chen C.Y., Margulies K.B., Shenoy V.B., and Prosser B.L.. 2016. Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science. 352:aaf0659. 10.1126/science.aaf0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu, J., Cade N.I., and Surrey T.. 2015. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 17:1422–1434. 10.1038/ncb3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz, C.A., Santarella R., Hoenger A., Karsenti E., Mattaj I.W., Gruss O.J., and Carazo-Salas R.E.. 2003. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 22:2060–2070. 10.1093/emboj/cdg195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgourdou, P., Mishra-Gorur K., Saotome I., Henagariu O., Tuysuz B., Campos C., Ishigame K., Giannikou K., Quon J.L., Sestan N., et al. 2017. Disruptions in asymmetric centrosome inheritance and WDR62-Aurora kinase B interactions in primary microcephaly. Sci. Rep. 7:43708. 10.1038/srep43708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., and Ross J.L.. 2012. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125:2561–2569. 10.1242/jcs.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S.C., Im S.K., Jang E.H., Jin K.S., Hur E.M., and Kim E.E.. 2019. Structural and Molecular Basis for Katanin-Mediated Severing of Glutamylated Microtubules. Cell Rep. 26:1357–1367.e5. 10.1016/j.celrep.2019.01.020 [DOI] [PubMed] [Google Scholar]
- Shohayeb, B., Lim N.R., Ho U., Xu Z., Dottori M., Quinn L., and Ng D.C.H.. 2018. The Role of WD40-Repeat Protein 62 (MCPH2) in Brain Growth: Diverse Molecular and Cellular Mechanisms Required for Cortical Development. Mol. Neurobiol. 55:5409–5424. 10.1007/s12035-017-0778-x [DOI] [PubMed] [Google Scholar]
- Thawani, A., Stone H.A., Shaevitz J.W., and Petry S.. 2019. Spatiotemporal organization of branched microtubule networks. elife. 8:e43890. 10.7554/eLife.43890 [DOI] [PMC free article] [PubMed]
- Vargas-Hurtado, D., Brault J.B., Piolot T., Leconte L., Da Silva N., Pennetier C., Baffet A., Marthiens V., and Basto R.. 2019. Differences in Mitotic Spindle Architecture in Mammalian Neural Stem Cells Influence Mitotic Accuracy during Brain Development. Curr. Biol. 29:2993–3005.e9. 10.1016/j.cub.2019.07.061 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., and Salmon E.D.. 1997. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 139:417–434. 10.1083/jcb.139.2.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek, M., Bechstedt S., Chaaban S., and Brouhard G.J.. 2015. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 17:907–916. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- Woodruff, J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., and Hyman A.A.. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 169:1066–1077.e10. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Xu, D., Zhang F., Wang Y., Sun Y., and Xu Z.. 2014. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Rep. 6:104–116. 10.1016/j.celrep.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Xu, D., Yao M., Wang Y., Yuan L., Hoeck J.D., Yu J., Liu L., Yeap Y.Y.C., Zhang W., Zhang F., et al. 2018. MEKK3 coordinates with FBW7 to regulate WDR62 stability and neurogenesis. PLoS Biol. 16:e2006613. 10.1371/journal.pbio.2006613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topçu M., McDonald M.T., Barry B.J., Felie J.M., et al. 2010. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 42:1015–1020. 10.1038/ng.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Yang S.L., Yang M., Herrlinger S., Shao Q., Collar J.L., Fierro E., Shi Y., Liu A., Lu H., et al. 2019. Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 10:2612. 10.1038/s41467-019-10497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Qin Y., Qin Y., Xu B., Guo T., Ke H., Chen M., Zhang L., Han F., Li Y., et al. 2018. Wdr62 is involved in female meiotic initiation via activating JNK signaling and associated with POI in humans. PLoS Genet. 14:e1007463. 10.1371/journal.pgen.1007463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents CRISPR-Cas9 gRNA targeting sequences and PCR primers for generating homology recombination donor constructs and genotyping.