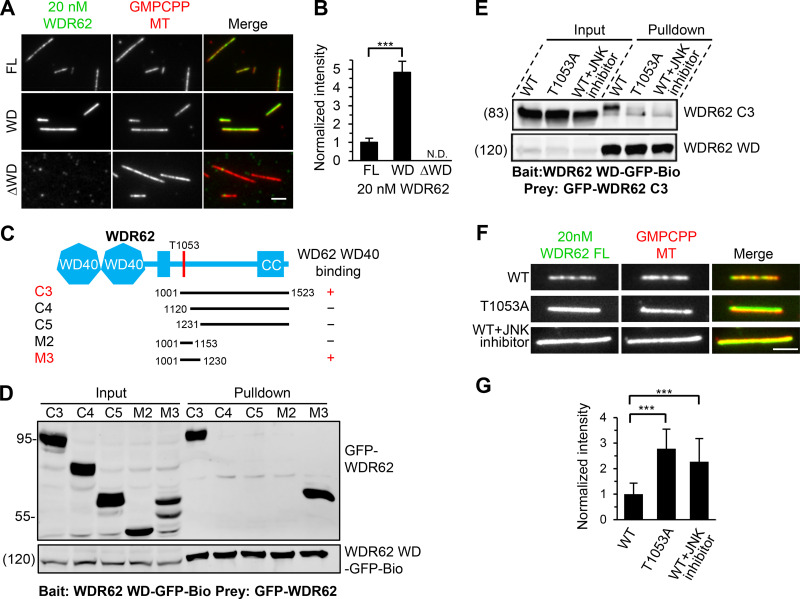
Figure 5.
The MT-binding affinity of WDR62 is autoinhibited through JNK phosphorylation–induced intramolecular interaction.(A and B) Images and quantification of the binding of 20 nM GFP-tagged WDR62 FL or its indicated truncations (green) to GMPCPP-stabilized MTs (red). The values were normalized to the intensity of the WDR62 FL-GFP. N.D., not detected. For all conditions, n = 60 MTs from two experiments. (C) Schematic overview of the domain organization of WDR62 and the deletion mutants and summary of their interactions with WD40 domain. CC, coiled-coil. (D) Streptavidin pull-down assays with extracts of HEK293T cells expressing WDR62-WD-GFP-Bio (bait) together with the indicated GFP-tagged WDR62 truncations (prey) analyzed by Western blotting with GFP antibody. (E) Streptavidin pull-down assays with extracts of HEK293T cells expressing WDR62-WD-GFP-Bio (bait) together with GFP-tagged WT WDR62 C3 fragment with or without JNK inhibitor treatment or its T1053A mutant (prey) analyzed by Western blotting with GFP antibody. (F and G) Images and quantification of the binding of 20 nM GFP-tagged FL WDR62 WT with or without JNK inhibitor treatment or T1053A mutant (green) to GMPCPP-stabilized MTs (red). The values were normalized to the intensity of the WT WDR62-GFP without JNK inhibitor treatment. From left to right, n = 150, 150, and 124 MTs from three experiments. Scale bars, 2 µm. Data represent mean ± SD. ∗∗∗, P < 0.001; two-tailed t test.