Guerreiro et al. show that loss of the primary microcephaly gene WDR62 in human cells mislocalizes the microtubule-severing enzyme katanin from mitotic spindle poles. This mislocalization reduces microtubule minus-end depolymerization, slowing poleward microtubule flux and promoting asynchronous chromosome segregation in anaphase.
Abstract
Mutations in the WDR62 gene cause primary microcephaly, a pathological condition often associated with defective cell division that results in severe brain developmental defects. The precise function and localization of WDR62 within the mitotic spindle is, however, still under debate, as it has been proposed to act either at centrosomes or on the mitotic spindle. Here we explored the cellular functions of WDR62 in human epithelial cell lines using both short-term siRNA protein depletions and long-term CRISPR/Cas9 gene knockouts. We demonstrate that WDR62 localizes at spindle poles, promoting the recruitment of the microtubule-severing enzyme katanin. Depletion or loss of WDR62 stabilizes spindle microtubules due to insufficient microtubule minus-end depolymerization but does not affect plus-end microtubule dynamics. During chromosome segregation, WDR62 and katanin promote efficient poleward microtubule flux and favor the synchronicity of poleward movements in anaphase to prevent lagging chromosomes. We speculate that these lagging chromosomes might be linked to developmental defects in primary microcephaly.
Introduction
The goal of the mitotic spindle is to faithfully segregate sister chromatids into two daughter cells (Walczak et al., 2010). The bipolar spindle relies on dynamic microtubules to attach and align sister-chromatids on the metaphase plate, before pulling them apart toward each spindle pole during anaphase (Prosser and Pelletier, 2017). Microtubules are polar polymers composed of tubulin dimers, with minus-ends embedded into spindle poles and plus-ends attached to kinetochores. Microtubules are dynamic, changing in a stochastic manner between growth and shrinkage, allowing them to explore their 3D environment (Akhmanova and Steinmetz, 2015; Prosser and Pelletier, 2017). Once microtubules bind kinetochores, they become more stable and form bundles called kinetochore fibers (k-fibers). These fibers still remain dynamic, as their plus-ends undergo periods of relative growth and shrinkage, resulting in oscillatory movements of bipolarly attached sister-kinetochores along the spindle axis (Jaqaman et al., 2010; Skibbens et al., 1993). At the same time, microtubules within k-fibers undergo a conveyor belt–like movement called poleward microtubule flux that is driven by microtubule motors acting on kinetochore and nonkinetochore microtubules, which over time results in new tubulin dimer incorporation at their plus-ends, concomitant with tubulin dimer removal at minus-ends (Mitchison, 1989; Steblyanko et al., 2020). The entire segregation machinery is controlled by the spindle assembly checkpoint, which ensures that cells initiate anaphase only once all kinetochores are attached to spindle microtubules (Joglekar, 2016).
Mitotic spindle defects are associated with severe human pathologies. In cancer, errors in chromosome segregation are frequent and contribute to high genetic instability (Levine and Holland, 2018). The most frequent cause for the gain or loss of entire chromosomes is the presence of merotelic kinetochore–microtubule attachments (one sister-kinetochore bound by microtubules of both poles; Gregan et al., 2011). Such erroneous attachments arise in a stochastic manner, but persist when microtubule dynamics are perturbed, preventing their correction (Bakhoum et al., 2009a; Godek et al., 2015). They can also accumulate after transient spindle defects, such as the formation of monopolar, multipolar, or bipolar spindles with incorrect geometry (Kaseda et al., 2012; McHedlishvili et al., 2012; Nam and van Deursen, 2014; Silkworth et al., 2012; 2009; Ganem et al., 2009). Autosomal recessive primary microcephaly (microcephaly primary hereditary [MCPH]) is the second major disease linked to genetic defects in the chromosome segregation machinery (Jayaraman et al., 2018; Nano and Basto, 2017). This developmental disease leads to severe underdevelopment of the neocortex, resulting in a small brain and mental retardation. Many MCPH mutations result in premature stop codons or low expression of genes encoding mitotic spindle components. These include proteins at centrosomes, spindle poles or kinetochores, and components of the DNA-damage pathway. Centrosomes are the main microtubule organizing centers of metazoan cells. They are composed of two barrel-shaped centrioles surrounded by pericentriolar material that acts as primary microtubule nucleation center during mitosis (Conduit et al., 2015). Although centrosomes are nonessential for cell division, they increase the fidelity of chromosome segregation, focus spindle poles, regulate spindle microtubule dynamics, and act as anchors for spindle orientation and positioning during asymmetric cell divisions (Basto et al., 2006; Meraldi, 2016; Sir et al., 2013; Dudka et al., 2019).
The cellular origins for primary microcephaly are most likely diverse. They include spindle orientation defects, chromosome segregation errors and aneuploidy, or accumulation of DNA damage (Jayaraman et al., 2018; Nano and Basto, 2017). Their common end point is exhaustion of dividing neuronal stem cells, either because they differentiate too early or because they undergo apoptosis. One important question is why such general cell cycle defects would mostly lead to brain developmental defects (Woods and Basto, 2014). One possibility is that cell fate determination is particularly sensitive to spindle orientation defects and/or that neuronal stem cells are particularly prone to apoptosis in the presence of aneuploidy or DNA damage (Gogendeau et al., 2015; Lancaster and Knoblich, 2012; Nano and Basto, 2017; Noatynska et al., 2012). A second key question is how mutations in different types of centrosomal proteins lead to primary microcephaly. A first large class of mutants is linked to genes required for centrosome duplication (SAS6, CPAP, CEP63, CEP152, CEP135, STIL, and CDKRAP2/CEP215; Nigg et al., 2014; O’Neill et al., 2018), which can impair the entire centrosome. A second class of mutations is found in genes involved in spindle pole organization, including ASPM, the microtubule-severing enzyme complex katanin, CDK5RAP2, CDK6, and WDR62 (Bilgüvar et al., 2010; Bond et al., 2002; 2005; Hu et al., 2014; Hussain et al., 2013; Mishra-Gorur et al., 2014; Nicholas et al., 2010; Yu et al., 2010). WDR62 (also called MCPH2), is the second most frequently mutated gene associated with primary microcephaly. The precise cellular function of the WDR62 protein and its localization remain under debate. WDR62 has been proposed to be associated with spindle poles, centrosomes, and/or spindle microtubules (Jayaraman et al., 2016; Kodani et al., 2015; Lim et al., 2016; Yu et al., 2010; Lim et al., 2015). Its localization is controlled by the protein kinase aurora-A, which phosphorylates WDR62 at mitotic onset to recruit it at centrosomes/spindle poles (Chen et al., 2014; Lim et al., 2016, 2015). At the functional level, human or murine WDR62 has been implicated in centriole duplication in conjunction with ASPM (Jayaraman et al., 2016), cilia formation when interacting with CPAP/IFT88 (Shohayeb et al., 2020), and cilia disassembly when interacting with CEP170 and KIF2A (Zhang et al., 2019). Loss of WDR62 has been reported to affect spindle orientation (Bogoyevitch et al., 2012; Miyamoto et al., 2017), spindle formation and mitotic progression without spindle orientation defects (Chen et al., 2014), or centrosome duplication and cilia formation (Jayaraman et al., 2016).
Here, we investigated the cellular functions of WDR62 in malignant and nontransformed human epithelial cell lines using both short-term siRNA depletions and long-term CRISPR/Cas9 gene knockouts. Our results indicate that WDR62 localizes to microtubule ends at spindle poles, but that it is not associated with centrosomes. We demonstrate that WDR62 and the microtubule-severing enzyme katanin are mutually dependent to localize at spindle poles. Loss of WDR62 stabilizes spindle microtubules due to insufficient microtubule minus-end depolymerization, while leaving plus-end microtubule dynamics unaffected. At the level of the spindle, WDR62 loss leads to a severe reduction of poleward microtubule flux in metaphase, concomitant with asynchronous poleward movements in anaphase that result in lagging chromosomes. We propose that these lagging chromosomes could be linked to developmental defects in primary microcephaly.
Results
WDR62 localizes to the microtubules at spindle poles
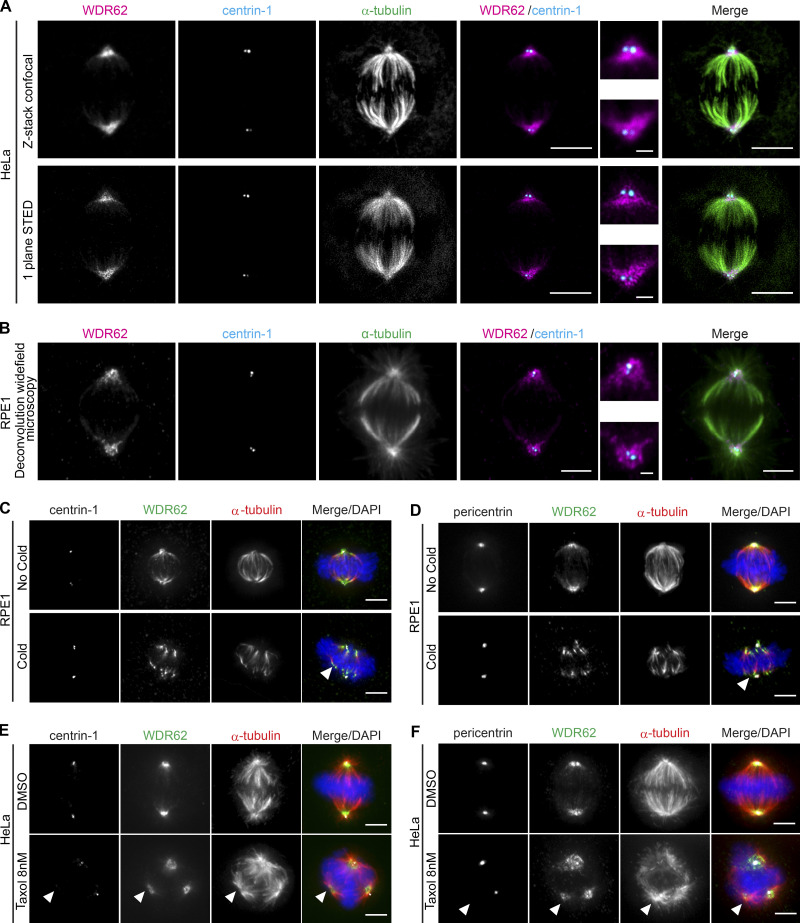
To study the function of WDR62 in mitosis, we first investigated its precise localization on the mitotic spindle. Previous studies reported that it binds to centrosomes (Jayaraman et al., 2016; Kodani et al., 2015; Yu et al., 2010), while others suggested a potential binding to microtubules (Lim et al., 2016; 2015). To localize WDR62, we used both malignant HeLa cells and nonmalignant human retinal pigment epithelial cells expressing telomerase (hTert-RPE1), two cell lines widely used for mitotic studies. Cells were stained for centrin-1 (centriole marker), α-tubulin (microtubule marker), WDR62, and DAPI (chromosome marker). Using both deconvolution wide-field microscopy (RPE1) and 3D stimulated emission depletion (STED) superresolution microscopy (HeLa), we found that WDR62 was not enriched at centrioles, but rather localized to spindle poles, possibly as part of the pericentriolar material, or in association with the microtubules terminating at spindle poles (Fig. 1, A and B).
Figure 1.
WDR62 localizes to microtubules at spindle poles.(A and B) Immunofluorescence images of metaphase. HeLa cells recorded by confocal (z-stack) and STED (one-plane) microscopy (A) or RPE1 cells recorded by deconvolution wide-field microscopy (one plane; B); cells were stained with α-tubulin, centrin-1, and WDR62 antibodies. Insets show spindle poles. (C and D) Immunofluorescence images of metaphase RPE1 cells, treated with or without a cold treatment and stained with DAPI, WDR62 and α-tubulin antibodies, and centrin-1 (C) or pericentrin (D) antibodies. Arrows indicate depolymerized microtubule minus-ends. (E and F) Immunofluorescence images of metaphase HeLa cells treated either with DMSO or 8 nM taxol for 12 h, stained with DAPI, WDR62 and α-tubulin antibodies, and centrin-1 (E) or pericentrin (F) antibodies. Arrowheads indicate centriole- and pericentrin-free spindle poles. Scale bars = 5 µm; 1 µm (inset).
To differentiate between the two possibilities, RPE1 cells were submitted to an ice-cold treatment for 6 min. Such treatment depolymerizes free microtubules and partially depolymerizes spindle microtubules, resulting in their detachment from the pericentriolar material. We found that WDR62 was exclusively bound to the dissociated spindle microtubules (Fig. 1 C). In contrast, the pericentriolar marker pericentrin was still associated with centrosomes (Fig. 1 D). This suggested that WDR62 does not bind centrosomes but is rather associated with microtubule ends terminating at spindle poles.
To confirm this hypothesis, we used 8 nM of the microtubule-stabilizing drug taxol on HeLa cells to form multipolar spindles with centrosome-free and centrosome-associated spindle poles (Fig. 1 E; Jordan et al., 1993). Immunofluorescence analysis confirmed that WDR62, but not pericentrin, was present in the extra spindle poles free of centrosomes and centrioles (Fig. 1, E and F). We conclude that WDR62 is located at microtubule ends terminating at spindle poles, independently of centrosomes.
Loss of WDR62 increases spindle microtubule stability
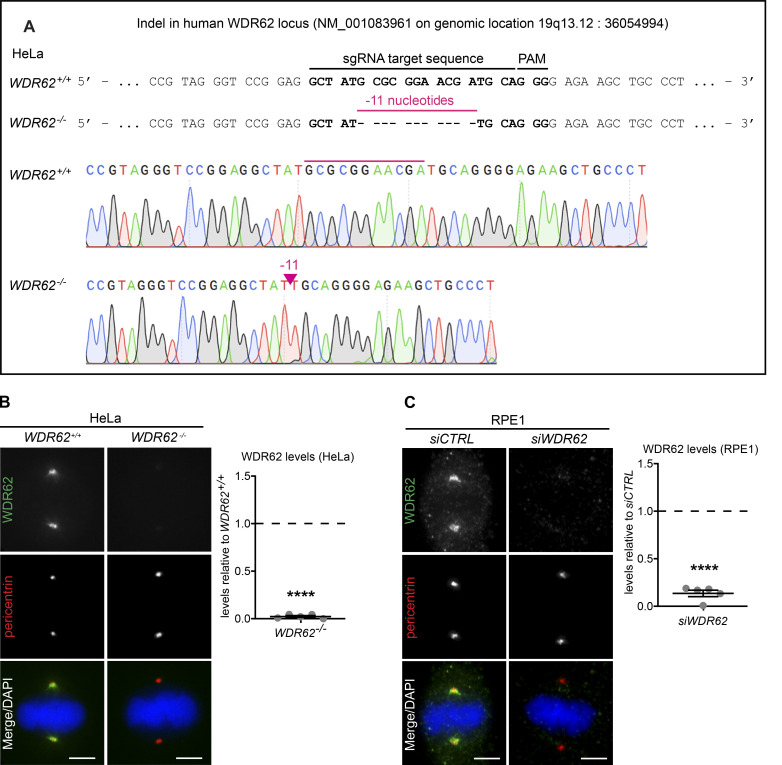
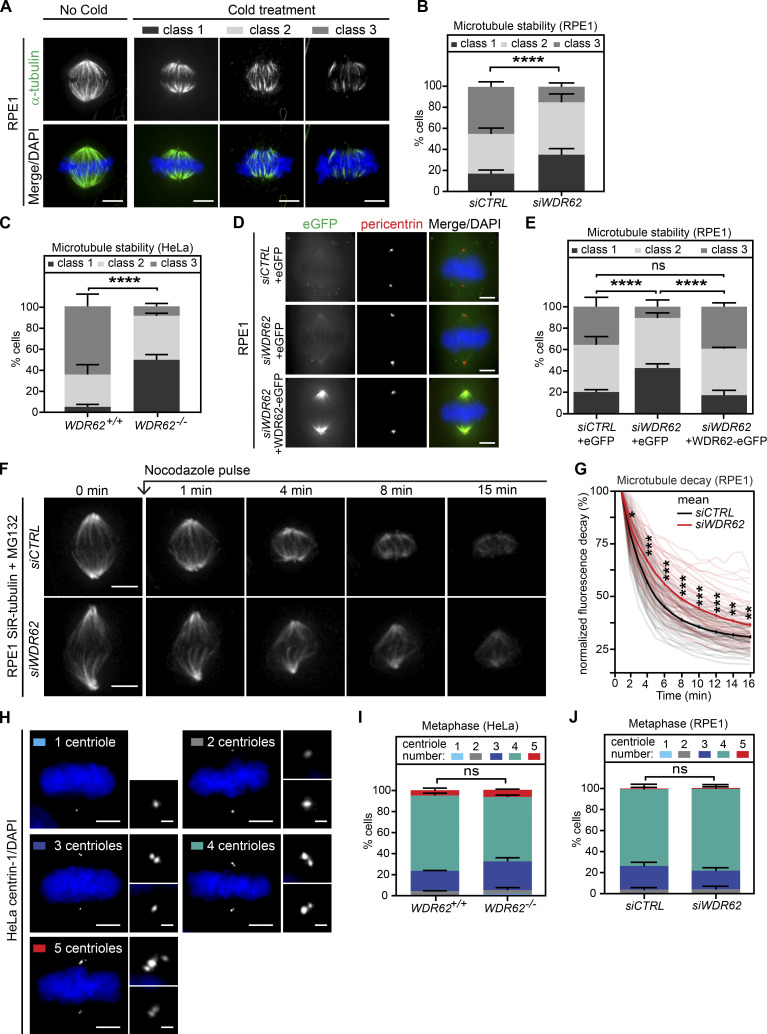
Given the localization of WDR62 on microtubules at spindle poles, we tested how WDR62 loss affects spindle microtubule dynamics. Previous studies had linked WDR62 to microtubule stability, reporting that it could promote astral microtubule assembly (Miyamoto et al., 2017) and drive the disassembly of microtubules in the cilium via the microtubule depolymerase KIF2A (Zhang et al., 2019). To probe WDR62 function in human cells, we either knocked out the WDR62 gene by CRISPR/Cas9 in HeLa cells expressing the chromosome marker H2B-GFP (WDR62−/−; genomic sequencing confirmed the presence of an 11-base deletion in the first exon, Fig. S1 A) or depleted it for 48 h by siRNA (siWDR62) in RPE1 cells. Immunofluorescence indicated that these approaches led to an ∼90% depletion (siWDR62) or a complete loss of WDR62 (WDR62−/−; Fig. S1, B and C). We subjected cells with or without WDR62 to ice-cold treatment (6 min for RPE1; 20 min for HeLa) to reveal the cold resistance of metaphase k-fibers. This resistance, in turn, reflects kinetochore-microtubule stability. Cells were classified into three categories based on 3D visualization after α-tubulin staining: an intact metaphase spindle with almost all k-fibers present (class 1), a metaphase spindle with several missing and detached k-fibers at microtubule minus- or plus-ends (class 2), and a deteriorated spindle with most k-fibers lost (class 3; Fig. 2 A). Both WDR62 depletion in RPE1 cells and WDR62 deletion in HeLa cells increased k-fiber cold resistance in metaphase when compared with control depletion or the parental cell line (Fig. 2, B and C). To exclude off-target effects, we created stable RPE1 cells expressing eGFP or an siRNA-resistant exogenous WDR62-eGFP that localized correctly at spindle poles (Fig. 2 D). RNAi-resistant WDR62-eGFP, but not eGFP alone, rescued cold sensitivity after siWDR62 treatment, suggesting that WDR62 is required for k-fiber destabilization (Fig. 2 E).
Figure S1.
WDR62 deletion and depletion efficiency. Related to Fig. 2. (A) Genomic sequence before and after performing CRISPR-Cas9 WDR62 knockout on HeLa H2B-GFP cells. PAM indicates the protospacer adjacent motif GGG. (B) Immunofluorescence images of metaphase WDR62+/+ and WDR62−/− HeLa H2B-GFP cells, stained with WDR62 and pericentrin antibodies and DAPI (left); quantification of WDR62 levels relative to WDR62+/+ (right): N = 5, n = 310 (WDR62+/+) and 322 (WDR62−/−) cells; dot plot represents median per experiment; bars represent mean ± SEM; ****, P < 0.0001, two-tailed unpaired t test. (C) Immunofluorescence images of metaphase RPE1 cells treated with siCTRL and siWDR62, stained with WDR62 and pericentrin antibodies and DAPI (left); quantification of WDR62 levels after siWDR62 treatment relative to siCTRL (right): N = 5, n = 310 cells per conditions; dot plot represents median per experiment; bars represent mean ± SEM; ****, P < 0.0001, two-tailed paired t test. All scale bars = 5 µm.
Figure 2.
WDR62 loss increases spindle microtubule stability.(A) Immunofluorescence images of cold-treated metaphase RPE1 cells, stained with α-tubulin antibodies and DAPI. Cells were categorized into three representative classes based on k-fiber integrity in 3D. (B and C) Quantification of cold-stable assay in metaphase RPE1 cells treated with siCTRL or siWDR62 (B) or metaphase HeLa WDR62+/+ or WDR62−/− cells (C); stack bars indicate mean percentages of class 1, 2, and 3 spindles as depicted in A: N = 3 independent experiments, n = 184–188 cells per condition (B) or n = 207–215 cells (C); error bars represent mean ± SEM; ****, P < 0.0001, χ2 test. (D) Immunofluorescence images of RPE1 cells expressing eGFP or WDR62-eGFP treated with siCTRL or siWDR62 and stained with pericentrin antibodies and DAPI. (E) Quantification of cold-stable assay in RPE1 eGFP or WDR62-eGFP metaphase cells treated with siCTRL or siWDR62 as in B and C: N = 3, n = 183–206 cells; ****, P < 0.0001, χ2 test. (F) Time-lapse sequences of MG132-arrested RPE1 metaphase cells treated with siCTRL or siWDR62, labeled with SiR-tubulin, and treated with 200 ng/ml nocodazole at t = 0. (G) Quantification of the spindle microtubule decay over time in RPE1 metaphase cells treated with siCTRL or siWDR62: N = 3, n = 55–58 cells; thick lines represent mean; thin lines represent single experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001, repeated two-tailed paired t test. (H) Immunofluorescence images of metaphase HeLa cells stained with centrin-1 antibodies and DAPI. Cells were categorized according to centriole numbers. (I and J) Quantification of centriole numbers in metaphase WDR62+/+ or WDR62−/− HeLa cells (I) or RPE1 cells treated with siCTRL or siWDR62 (J): stack bars indicate mean percentage of cells with corresponding centrioles numbers; error bars represent mean ± SEM; N = 1, n = 200 cells; χ2 test. Scale bars = 5 µm; 2.5 µm (inset).
To corroborate these findings, we used an unbiased spindle stability quantification assay in RPE1 cells. To visualize k-fibers, cells were labeled with SiR-tubulin, a tubulin live dye that binds to microtubules with a turnover rate of >10 s (Lukinavičius et al., 2014; David et al., 2019), at a concentration that does not change k-fiber dynamics (Dudka et al., 2019). Cells were arrested in metaphase with the proteasome inhibitor MG132 and treated with a 200 ng/ml pulse of the microtubule-depolymerizing drug nocodazole. Spindle decay was monitored live over 15 min, as previously described (Fig. 2 F; Dudka et al., 2019; Wilhelm et al., 2019). This assay indicated that k-fibers were significantly more stable after WDR62 depletion (Fig. 2 G), consistent with the cold treatment results.
Previous studies had reported that WDR62 is necessary for efficient centrosome duplication (Jayaraman et al., 2016; Kodani et al., 2015). We therefore tested, in both HeLa WDR62−/− and RPE1 cells treated with control or WDR62 siRNA, whether the change in microtubule stability could be due to impaired centrosome duplication. By counting the number of centrioles in metaphase with centrin-1 antibodies, we found that WDR62 loss or acute depletion was not associated with a significant centriole loss (Fig. 2, H–J). This indicates that WDR62 is either not involved in centrosome duplication or that it plays only a minor role in this process. We conclude that it promotes k-fiber destabilization independently of centriole numbers.
WDR62 is required for katanin localization at spindle poles
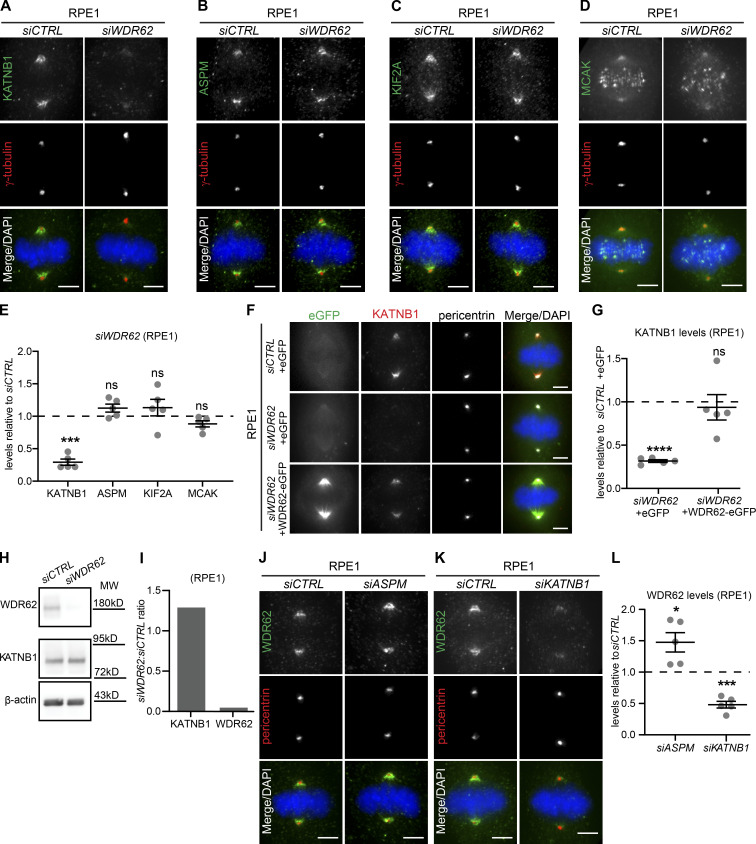
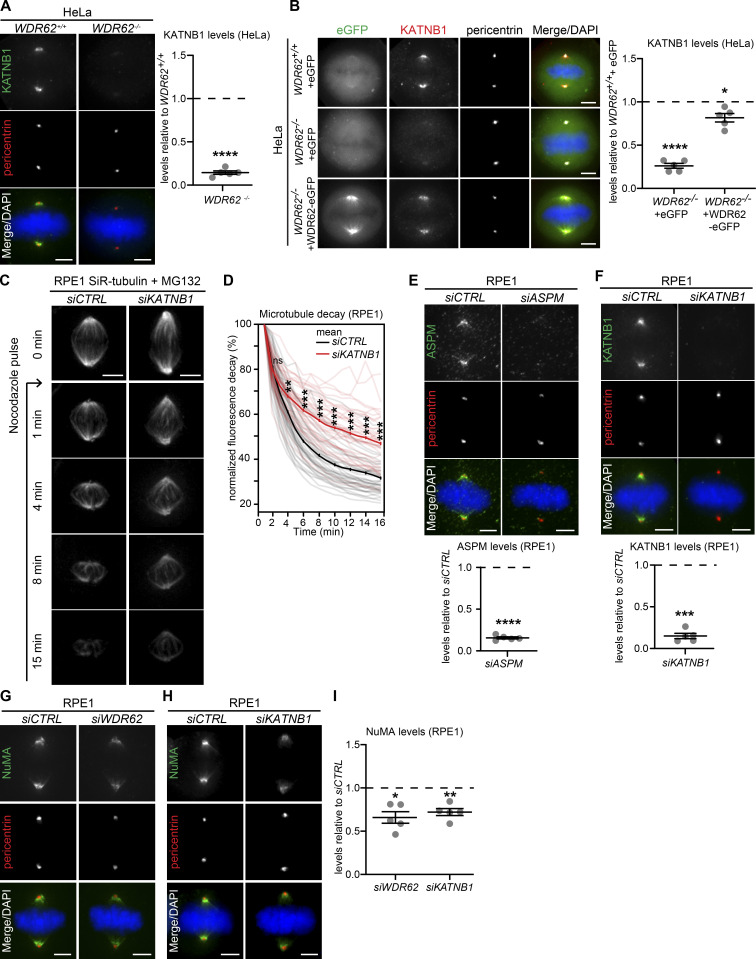
WDR62 has been shown to interact with two different regulators of microtubule stability: the microtubule-depolymerase KIF2A at cilia (Zhang et al., 2019) and ASPM (MCPH5), another microcephaly-associated gene product, whose presence was reported to promote the localization of the microtubule-severing enzyme katanin (KATNB1) at spindle poles (Bond et al., 2002; 2003; Jayaraman et al., 2016; Jiang et al., 2017). We immunostained control- and siWDR62-treated cells for katanin, ASPM, KIF2A, and MCAK (a KIF2A paralog that acts at spindle poles and kinetochores) and quantified their levels at spindle poles. WDR62 depletion did not alter ASPM, KIF2A, or MCAK levels at spindle poles; katanin levels, however, were decreased by >70% (Fig. 3, A–E). Katanin levels at spindle poles were also dramatically reduced in WDR62−/− HeLa cells (−74%; Fig. S2 A) and could be rescued with siRNA-resistant WDR62-eGFP in siWDR62-treated RPE1 cells (Fig. 3, F and G) or with exogenous WDR62-eGFP in HeLa WDR62−/− cells (Fig. S2 B). This demonstrated that the katanin decrease at spindle poles is not an off-target effect. Immunoblotting indicated that WDR62 depletion in RPE1 cells did not affect global KATNB1 levels (Fig. 3, H and I), but only prevented its correct localization at spindle poles. In line with these results, KATNB1 depletion stabilized spindle microtubules even more than WDR62 depletion in the spindle decay assay (Fig. S2, C and D).
Figure 3.
WDR62 is required for katanin localization at spindle poles.(A–D) Immunofluorescence images of metaphase RPE1 cells treated with siCTRL or siWDR62, stained with DAPI, antibodies against γ-tubulin, and KATNB1 (A), ASPM (B), KIF2A (C), or MCAK (D). (E) Quantification of protein levels at spindle poles in siWDR62 relative to siCTRL: N = 5; n = 286–328 cells; dot plot represents median per experiment; bars represent mean ± SEM; ***, P < 0.001, two-tailed paired t test. (F) Immunofluorescence images of RPE1 eGFP or WDR62-eGFP metaphase cells transfected with indicated siRNA, stained with KATNB1 and pericentrin antibodies, and DAPI. (G) Quantification of KATNB1 levels at spindle poles as shown in F: N = 5, n = 263–279 cells; dot plot represents median per experiment; bars represent mean ± SEM; ****, P < 0.0001, one-way ANOVA. (H) Western blot of asynchronous RPE1 cell lysates treated with siCTRL or siWDR62 and probed against WDR62, KATNB1, and β-actin (loading control). (I) Quantification of immunoblot shown in H of KATNB1/β-actin and WDR62/β-actin ratio signal in siWDR62 relative to siCTRL. N = 1. (J and K) Immunofluorescence images of metaphase RPE1 cells treated with indicated siRNA, stained with WDR62, pericentrin antibodies, and DAPI. (L) Quantifications of WDR62 levels at spindle poles in siASPM and siKATNB1 relative to siCTRL: N = 5; n = 320–339 cells; dot plot represents median per experiment; bars represent mean ± SEM; *, P < 0.05; ***, P < 0.001, two-tailed paired t test. All scale bars = 5 µm.
Figure S2.
KATNB1 depletion mimics WDR62 depletion. Related to Fig. 3. (A) Immunofluorescence images of metaphase WDR62+/+ and WDR62−/− HeLa cells, stained with KATNB1 and pericentrin antibodies and DAPI (left); quantification of KATNB1 levels in WDR62−/− relative to WDR62+/+(right): N = 5, n = 308 (WDR62+/+) and 292 (WDR62−/−) cells; dot plot represents median per experiments; bars represent mean ± SEM; ****, P < 0.0001, two-tailed unpaired t test. (B) Immunofluorescence images of metaphase WDR62+/+ or WDR62−/− HeLa cells transfected with indicated constructs and stained with KATNB1 and pericentrin antibodies and DAPI (left). Quantification of KATNB1 levels at spindle poles (right) in N = 5, n = 217–228 cells; dot plot represents median per experiment; bars represent mean ± SEM; *, P ≤ 0.05; ****, P < 0.0001, one-way ANOVA. (C) Time-lapse images of MG132-arrested RPE1 cells treated with siCTRL and siKATNB1, labeled with SiR-tubulin, and treated with 200 ng/ml of nocodazole at t = 0. (D) Quantification of the spindle microtubule decay over time in siCTRL- and siWDR62-treated RPE1 metaphase cells: N = 4, n = 50 (siCTRL) and 55 (siKATNB1) cells; thick line represents mean; thin line represents single experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001, repeated two-tailed paired t test. (E–I) Immunofluorescence of metaphase RPE1 cells treated with indicated siRNAs, stained with DAPI and antibodies against pericentrin and ASPM (E), KATNB1 (F), or NuMA (G and H); all quantifications (E and F below, and I right) represent the levels in the indicated siRNA vs. siCTRL: N = 5, n = 303–323 cells; dot plots represent median per experiments; bars represent mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, in two-tailed paired t test. All scale bars = 5 µm.
We next tested whether, conversely, ASPM or KATNB1 depletion would impact WDR62 abundance at spindle poles (Fig. 3, J–L). Both siRNA treatments efficiently depleted either protein within 48 h (Fig. S2, E and F). Quantitative analysis indicated that ASPM depletion led to a 48% increase in WDR62 levels at spindle poles (Fig. 3, J and L). We conclude that WDR62 does not require ASPM to localize to spindle poles. In contrast, KATNB1 depletion decreased WDR62 levels at spindle poles by 52% (Fig. 3, K and L). This result had two potential explanations: (1) either KATNB1 and WDR62 are mutually dependent on each other for their recruitment at spindle poles, or (2) this reduction reflects a reduced number of microtubule minus-ends at spindle poles (each katanin cut generates an additional minus-end). To differentiate between the two, we stained KATNB1- and WDR62-depleted cells for NuMA, a minus-end marker (Elting et al., 2014; Sikirzhytski et al., 2014; Gordon et al., 2001), and found a reduction of 30% and 34% in NuMA abundance at spindle poles (Fig. S2, G–I). Although we cannot exclude that WDR62 also specifically regulates NuMA recruitment, the results indicate that the strong reduction of WDR62 and KATNB1 in siKATNB1- or siWDR62-treated cells is specific, as it goes beyond the reduction seen with the minus-end marker NuMA. We conclude that WDR62 and katanin mutually depend on each other for spindle pole localization.
WDR62 loss impairs microtubule minus-end depolymerization
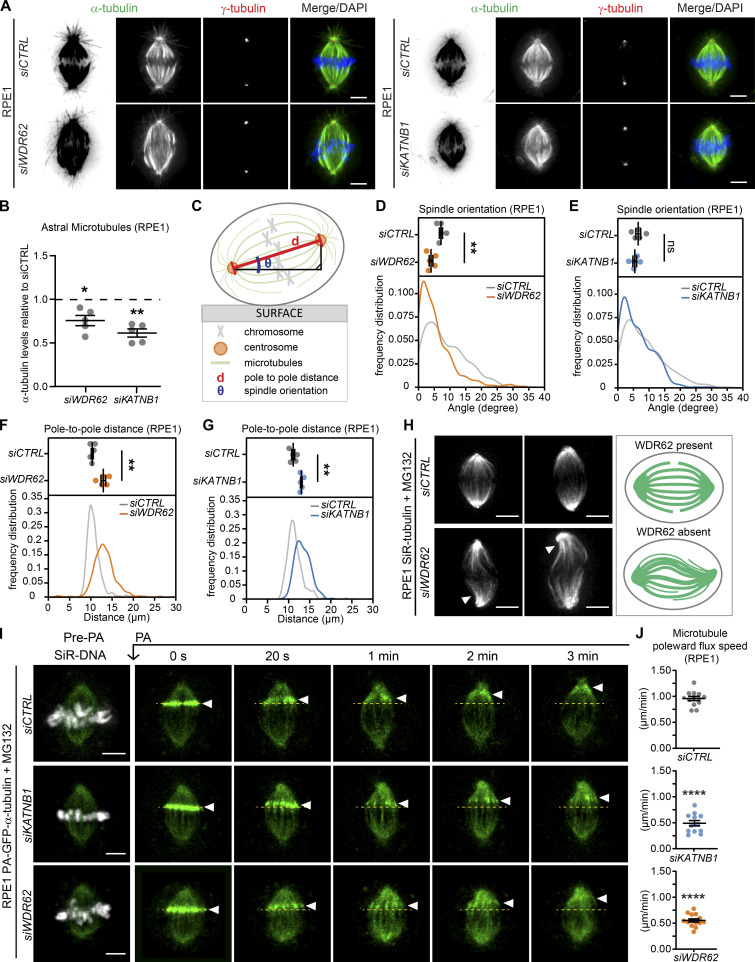
How does the loss of katanin at spindle poles affect the mitotic spindle? Some studies have reported that WDR62 is necessary for correct spindle orientation (Bogoyevitch et al., 2012; Hu and Jasper, 2019; Miyamoto et al., 2017; Ramdas Nair et al., 2016; Xu et al., 2014), while others did not observe such an effect (Chen et al., 2014; Jayaraman et al., 2016). Since spindle orientation depends on a correct interaction between astral microtubules and the cell cortex, we first analyzed whether WDR62 or KATNB1 depletion affects the density of astral microtubules. Our quantification of the α-tubulin signal behind the spindle poles indicated that both WDR62 and KATNB1 depletion led to 25–33% reduction in astral microtubule density (Fig. 4, A and B). This implied a modified balance in the distribution of microtubules in the spindle, with a partial reduction in astral microtubules and a higher k-fiber stability.
Figure 4.
WDR62 loss impairs microtubule minus-end depolymerization.(A) Immunofluorescence images of RPE1 cells treated with indicated siRNAs stained with antibodies against α- and γ-tubulin and DAPI. (B) Quantification of astral microtubules in siWDR62 and siKATNB1 relative to siCTRL; N = 5, n = 244–281 cells; dot plot represents median per experiment; bars represent mean ± SEM; *, P < 0.05; **, P < 0.01, two-tailed paired t test. (C) Scheme representing mitotic spindle orientation as the angle from the surface (θ) and the pole-to-pole distance (d). (D–G) Quantification of spindle orientation (D and E) and pole-to-pole distances (F and G) in siCTRL-, siKATNB1-, and siWDR62-treated RPE1 metaphase cells. Dot plots show medians per experiment; curves the frequency distribution; N = 5; n = 315–323 cells; bars represent mean ± SEM; **, P < 0.01 in two-tailed unpaired t test. (H) Immunofluorescence image of siCTRL- or siWDR62-treated MG132 arrested RPE1 cells labeled with SiR-tubulin, and representative spindle architecture scheme indicating microtubule buckling in the spindle of siWDR62-treated cells. (I) Time-lapse images of RPE1 PA-GFP-α-tubulin cells treated with indicated siRNAs and MG132, labeled with SiR-DNA, and photoactivated (PA) at t = 0. (J) Quantification of the microtubule poleward flux speed (μm/min): N = 3, n = 12–14 cells; dot plots represent each cell; bars represent mean ± SEM; ****, P < 0.0001, two-tailed paired t test. All scale bars = 5 µm. See also Video 1, Video 2, and Video 3.
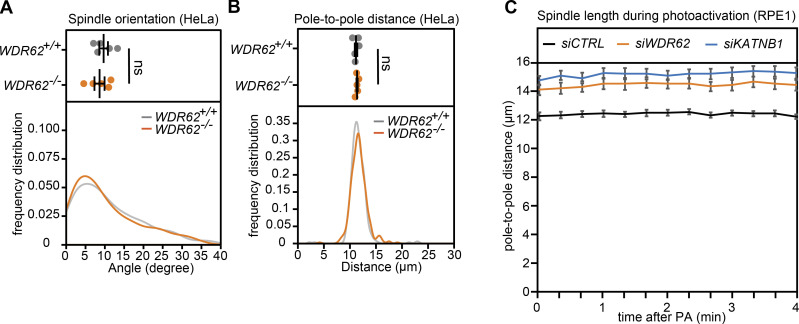
Next, we investigated the functional consequences of these changes, by monitoring spindle orientation. Since wild-type epithelial cells orient their spindle parallel to the growth surface, spindle orientation defects result in a higher median angle between the growth surface and the spindle axis (Fig. 4 C; Toyoshima and Nishida, 2007). However, both WDR62 and KATNB1 depletion in RPE1 cells did not increase the median spindle orientation angle, but if anything, led to a decrease in the angle (Fig. 4, D and E). The lack of spindle orientation defects was not due to incomplete protein depletion, since a similar phenotype was visible in HeLa WDR62−/−cells (Fig. S3 A). Both depletions, however, increased spindle length (Fig. 4, F and G), which could explain the reduced median spindle angle. Moreover, during the recording of the spindle decay assay, we often observed buckling of spindle microtubules between the two poles, implying a compression of microtubules as they push against the mitotic spindle (Fig. 4 H; Tolić et al., 2019). Overall, we conclude that neither WDR62 nor KATNB1 depletion results in a spindle orientation defect, but rather leads to buckling microtubules that arise due to the resistance of the mitotic spindle to outward pushing forces. This raised the hypothesis that spindles without WDR62 have an insufficient depolymerization at microtubule minus-ends, due to the absence of katanin.
Figure S3.
WDR62−/− HeLa cells show no difference in spindle orientation or pole-to-pole distance, and spindle length of MG132-arrested RPE1 cells are constant. Related to Fig. 4. (A and B) Quantification of spindle orientation (A) and pole-to-pole distances (B) in WDR62+/+ and WDR62−/− metaphase HeLa H2B-GFP cells; dot plots represent median per experiments with bars displaying mean ± SEM (top) and frequency distribution (bottom): N = 5, n = 323 (WDR62+/+) and 330 (WDR62−/−) cells; two-tailed unpaired t test. (C) Quantification of spindle length during photoactivation (PA) experiments in indicated siRNA treatments; lines represent mean per condition, with bars displaying SEM. n = 15 cells per condition.
To test this hypothesis, we quantified the rate of poleward microtubule flux in control-, WDR62-, and KATNB1-depleted RPE1 cells expressing photoactivatable (PA)-GFP-α-tubulin. This process is driven by microtubule motors in the spindle but requires microtubule minus-end depolymerization at spindle poles and new tubulin subunit incorporation at kinetochores (Buster et al., 2007; Mitchison, 1989; Steblyanko et al., 2020; Ganem et al., 2005). By photoactivating tubulin on k-fibers in metaphase cells arrested with the proteasome inhibitor MG132, when spindle length is constant (Fig. S3 C), one can read out the rate of minus-end depolymerization from the speed of poleward microtubule flux. Our quantification indicated that flux speeds were halved in WDR62- or KATNB1-depleted cells when compared with a control depletion (Fig. 4, I and J; and Videos 1, 2, and 3), indicating that KATNB1 loss at spindle poles reduces minus-end microtubule depolymerization by half.
Video 1.
Photoactivation of siCTRL-treated hTert-RPE1 PA-GFP-α-tubulin cells, showing microtubule poleward flux. Related to Fig. 4. Sampling rate = 20 s; scale bars = 5 µm. Play rate = 4 frames per second.
Video 2.
Photoactivation of siKATNB1-treated hTert-RPE1 PA-GFP-α-tubulin cells, showing microtubule poleward flux. Related to Fig. 4. Sampling rate = 20 s; scale bars = 5 µm. Play rate = 4 frames per second.
Video 3.
Photoactivation of siWDR62-treated hTert-RPE1 PA-GFP-α-tubulin cells, showing microtubule poleward flux. Related to Fig. 4. Sampling rate = 20 s; scale bars = 5 µm. Play rate = 4 frames per second.
WDR62 loss does not affect k-fiber plus-end dynamics but leads to wider metaphase plates
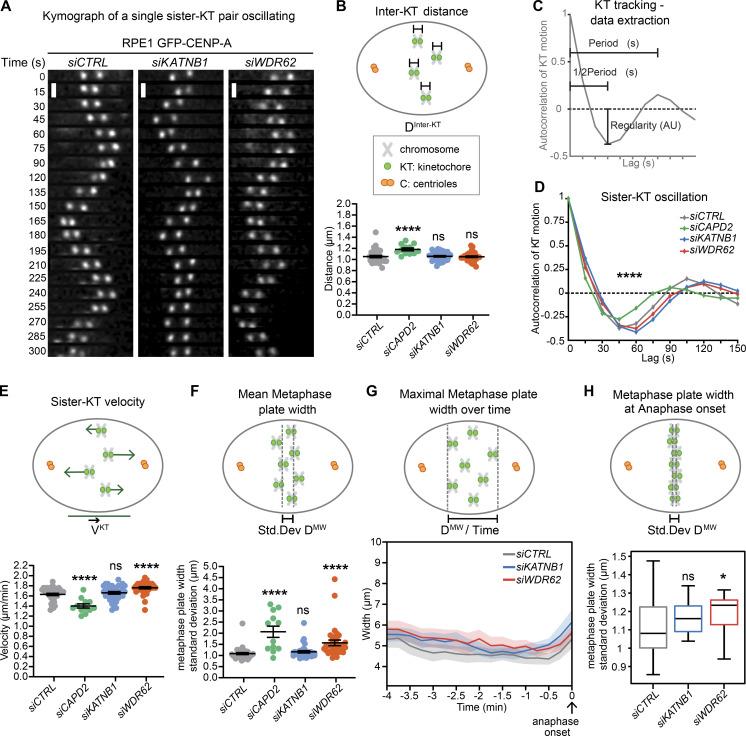
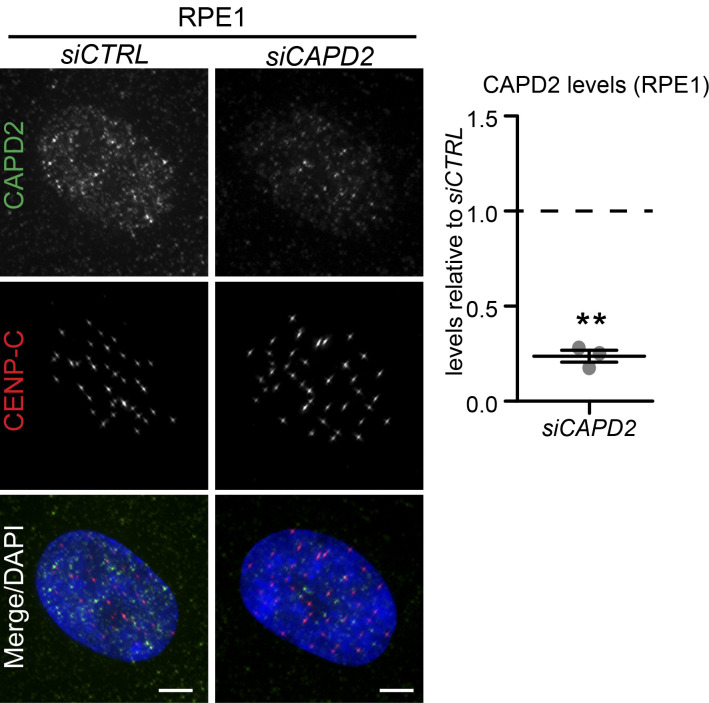
Having established that WDR62 depletion reduces k-fiber minus-end depolymerization, we tested whether it also influences their plus-ends and to which extent the dynamics of both ends are mechanically coupled. We used an automated kinetochore-tracking assay based on RPE1 cells expressing the kinetochore/centromere marker CENP-A (Jaqaman et al., 2010), as kinetochore movements on the metaphase plate reflect the dynamic instability of k-fiber plus-ends. Sister-kinetochores undergo semiregular oscillatory movements along the spindle axis, from which it is possible to extract key mechanochemical parameters of sister-kinetochores and k-fiber plus-ends (Fig. 5 A; Olziersky et al., 2018), as follows. (1) Interkinetochore distances, which depend on the sum of the forces pulling the two sister-kinetochores apart, and the mechanical stiffness of the centromeric DNA. WDR62 or KATNB1 depletion did not change interkinetochore distances; in contrast, depletion of the centromeric cohesin subunit CAPD2 increased interkinetochore distances, as previously observed (Figs. 5 B and S4 A; Jaqaman et al., 2010). (2) The autocorrelation of kinetochore movements reveals the average (half-)period and regularity of the oscillations (Fig. 5 C). These parameters reflect the dynamic instability of k-fibers, as the half-period indicates how frequently k-fibers switch between growth and shrinkage, while regularity indicates whether k-fibers behave in a consistent manner. WDR62 or KATNB1 depletion had at best only marginal effects on these parameters, while CAPD2 depletion led to shorter half-periods (Fig. 5 D). (3) The average speed of the kinetochore motions, which was slightly higher after WDR62 depletion, unchanged in KATNB1-depleted cells, and markedly decreased after CAPD2 depletion (Fig. 5 E). (4) The width of the metaphase plate, which reflects the ability of the spindle to balance the forces on sister-kinetochores and to align chromosomes on a tight metaphase plate as cells progress toward anaphase (Jaqaman et al., 2010). WDR62 depletion, as well as CAPD2 depletion (positive control), led to wider metaphase plates. (Fig. 5 F; Jaqaman et al., 2010). These wider plates persisted throughout metaphase until anaphase onset (Fig. 5, G and H). A similar trend could also be observed in katanin-depleted cells at the level of the maximal metaphase plate width and metaphase plate width at anaphase onset, but the difference was too small to be significant within our sample size (Fig. 5, G and H).
Figure 5.
WDR62 loss leaves k-fiber plus-end dynamics intact but leads to wider metaphase plates.(A–H) Automated sister-kinetochore tracking of RPE1 GFP-centrin1/GFP-CENPA cells treated with indicated siRNA. Shown are kymographs single, oscillating sister-kinetochores, scale bars = 1 µm (A); quantification of the interkinetochore distances (B); schematic illustrating the parameters of sister-kinetochore oscillation autocorrelation curves (C); autocorrelation curves of sister-kinetochore oscillations along spindle axis (D); sister-kinetochore velocities (E); mean metaphase plate width (F); maximal metaphase plate width over time (G); and mean metaphase plate width at anaphase onset (H). N = 3–5, n = 35 (siCTRL), 12 (siCAPD2), 34 (siKATNB1), and 33 (siWDR62) cells; dot plots represent each cell with bars displaying mean ± SEM; box plot represent mean ± SD; *, P < 0.05; ****, P < 0.0001, one-way ANOVA. AU, arbitrary units.
Figure S4.
Validation of CAPD2 depletion in RPE1 cells. Related to Fig. 5. Immunofluorescence images of interphase RPE1 cells treated with siCTRL and siCAPD2 and stained with CAPD2 and CENP-C antibodies and DAPI (left); quantification of CAPD2 levels in siCAPD2 relative to siCTRL (right): N = 3, n = 125 cells per condition; dot plot represents median per experiment; bars represent mean ± SEM; **, P < 0.01, two-tailed paired t test. Scale bars = 5 µm.
We conclude that reduced minus-end depolymerization at spindle poles has only very little effect on the dynamics of k-fiber plus-ends, indicating that the two extremities can be experimentally uncoupled from each other. Nevertheless, WDR62 depletion led to persistently wider metaphase plates, implying an impaired ability to position the chromosomes in the equatorial plane of the spindle.
WDR62 is required for synchronous poleward anaphase movements
Our results were reminiscent of models based on data from Drosophila melanogaster cells that postulated that poleward microtubule flux equalizes forces within the mitotic spindle and favors synchronous poleward anaphase movements (Matos et al., 2009). However, so far all the studies addressing the role of poleward microtubule flux in anaphase relied on knock-downs that affected both microtubule minus- and plus-end dynamics (such as the depletion of KIF2A and MCAK or the depletion of the plus-end binding protein CLASP and MCAK; Ganem et al., 2005; Matos et al., 2009). This prevented definite conclusions on the role of flux, since changes in plus-end microtubule dynamics affect the fidelity of chromosome segregation on their own (Bakhoum et al., 2009a; Godek et al., 2015). WDR62 depletion allows more precise experimental testing of the role of poleward microtubule flux in chromosome segregation, since it purely affects minus-end depolymerization.
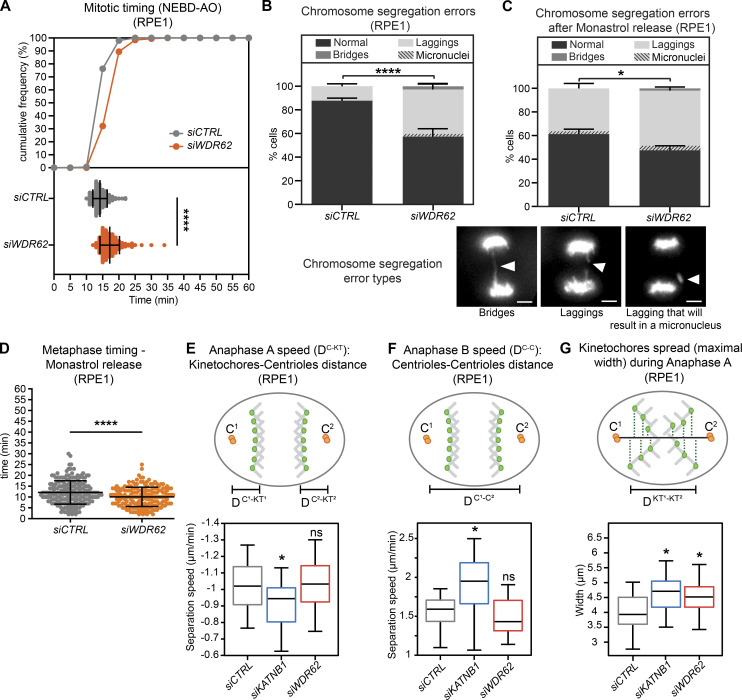
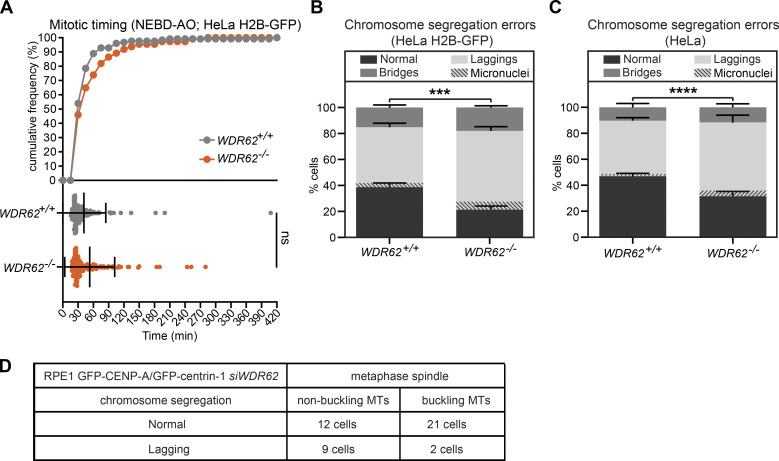
We first analyzed the overall behavior of control- and WDR62-depleted RPE1 cells expressing the chromosome marker H2B-GFP. Our results indicated that WDR62 depletion did not greatly affect the timing of chromosome segregation, as WDR62-depleted cells spent only 3 min more between nuclear envelope breakdown and anaphase (Fig. 6 A); nevertheless, WDR62 depletion led to a 22% increase in the number of cells displaying lagging chromosomes (35% vs. 13% after control depletion; Fisher’s exact test, P < 0.0001; Fig. 6 B and Videos 4, 5, and 6). In contrast, it did not significantly affect the percentage of chromosome bridges, which were rare in both conditions (Fisher’s exact test, P = 0.16; Fig. 6 B). The lagging chromosomes we observed in WDR62-depleted cells were very transient and only rarely led to the formation of micronuclei (Fig. 6 B). Since exogenous WDR62-GFP led to lagging chromosomes on its own (not depicted), we could not analyze the rescue experiment. Nevertheless, to test for off-target effects, we monitored chromosome segregation in HeLa or HeLa H2B-GFP, comparing the parental WDR62+/+ cells to WDR62−/− cells. While mitotic timing was unchanged after WDR62 loss (Fig. S5 A), the percentage of cells displaying lagging chromosomes was significantly higher in both knockouts (Fig. S5, B and C; P > 0.0001 and 0.0002 in χ2 test; note that the percentages of bridges were again not significantly changed, P = 0.34 and 0.24). The difference in the incidence of lagging chromosomes was smaller than in RPE1 cells (13% in both cases), most likely because of the already high level of lagging chromosomes in the parental HeLa cells. Since the same effect could be observed after siRNA depletion and two independent CRISPR/Cas9 knockouts in two different cell lines, we conclude that WDR62 loss leads to lagging chromosomes.
Figure 6.
WDR62 is required for synchronous poleward anaphase movements.(A) Mitotic timing from nuclear breakdown (NEBD) until anaphase onset (AO) of RPE1 EB3-GFP/H2B-mCherry cells treated with siCTRL or siWDR62, shown as cumulative frequency diagram or dot plot representing each cell with bars displaying mean ± SEM; N = 3, n = 156–203 cells; ****, P < 0.0001, two-tailed paired t test. (B and C) Percentage of chromosome segregation errors from RPE1 EB3-GFP/H2B-mCherry cells, either unsynchronized (B) or after monastrol release (C). Stack bars represent normal anaphase, anaphase with lagging chromosomes, or anaphase with chromosome bridges as shown on representative images; scale bars = 5 µm; gray stripes represent a cell with a lagging chromosome that will result in micronucleus; error bars represent mean ± SEM; N = 5, n = 125–209 cells for B; N = 3, n = 209–234 cells for C; ****, P < 0.0001; *, P = 0.042 by χ2 test. (D) Timing from monastrol arrest release until anaphase onset from C; dot plot represents each cell; bars represent mean ± SEM; ****, P < 0.0001, two-tailed paired t test. n = 44 cells. (E–G) Automated spindle pole and KT of RPE1 GFP-centrin1/GFP-CENPA cells treated with indicated siRNA. Shown are anaphase A speed, derived from the distance between kinetochores and centrioles (DC-KT) over time (E); anaphase B speed, derived from the intercentrosome distance (DC-C) over time (F); and the maximal spread of kinetochores during their movements toward the poles in anaphase A for each condition (G); N = 3, n = 20 cells; bars represent mean ± SD; *, P < 0.05, one-way ANOVA. See also Video 4, Video 5, Video 6, Video 7, and Video 8.
Video 4.
Mitotic progression of siCTRL-treated hTert-RPE1 EB3-GFP/H2B-mCherry cells, showing no error of chromosome segregation. Related to Fig. 6. Sampling rate = 1 min; scale bars = 5 µm. Play rate = 3 frames per second.
Video 5.
Mitotic progression of siWDR62-treated hTert-RPE1 EB3-GFP/H2B-mCherry cells, showing lagging chromosomes in anaphase. Related to Fig. 6. Sampling rate = 1 min; scale bars = 5 µm. Play rate = 3 frames per second.
Video 6.
Mitotic progression of siWDR62-treated hTert-RPE1 EB3-GFP/H2B-mCherry cells, showing DNA bridge in anaphase. Related to Fig. 6. Sampling rate = 1 min; scale bars = 5 µm. Play rate = 3 frames per second.
Figure S5.
Loss of WDR62 in HeLa cells does not affect mitotic timing but increases the percentage of lagging chromosomes. Related to Fig. 6. (A) Mitotic timing from nuclear breakdown (NEBD) until anaphase onset (AO) of WDR62+/+ and WDR62−/−HeLa H2B-GFP cells: cumulative frequency (top) with dot plot representing each cell (bottom); bars represent mean ± SEM; two-tailed unpaired t test. (B and C) Percentage of chromosome segregation errors from unsynchronized HeLa H2B-GFP (B) or HeLa (C) cells with a WDR62+/+ and WDR62−/− genotype. Stack bars indicate the number of normal anaphases, anaphases with lagging chromosomes, or anaphases with chromosome bridges; gray stripes represent cells with lagging chromosomes resulting in micronuclei; error bars represent mean ± SEM; N = 5, n = 290–367 cells; ***, P = 0.0002; ****, P < 0.0001, χ2 test. (D) Quantification of lagging chromosomes in SiR-tubulin–stained RPE1 GFP-centrin1/GFP-CENPA cells with buckling or nonbuckling microtubules after siWDR62 treatment.
In theory, lagging chromosomes can originate from an inefficient correction of merotelic kinetochore-microtubule attachments, possibly due to an overstabilization of k-fibers (Bakhoum et al., 2009b; Gregan et al., 2011; Thompson and Compton, 2008). To test for this hypothesis, we treated cells with the Eg5 inhibitor monastrol to achieve a monopolar spindle configuration, before releasing them from monastrol arrest. This configuration results in a high incidence of syntelic (both sister-kinetochores attached to the same pole) and merotelic attachments (Lampson et al., 2004). The time cells take to align the chromosome on the metaphase plate reflects their ability to correct syntelic attachment, while the percentage of lagging chromosomes reflects persistent merotelic attachments (Lampson et al., 2004). WDR62-depleted RPE1-H2B-GFP cells released from monastrol arrest aligned their chromosomes as rapidly as control-depleted cells and had an incidence of lagging chromosomes that was only marginally higher than control-depleted cells, and similar to that observed in cells not treated with monastrol (Fig. 6, C and D). This indicated that WDR62 does not have a major impact on the correction of erroneous kinetochore–microtubule attachments. A second possible explanation for the lagging chromosomes could be that buckling microtubules “push back” on segregating chromosomes. We therefore tested whether microtubule buckling in metaphase could be a predictor for lagging chromosomes in anaphase. Our analysis of live RPE1 eGFP-CENP-A/eGFP-centrin-1 stained with SiR-tubulin and depleted of WDR62, however, indicated that microtubule buckling in metaphase did not correlate with lagging chromosomes, rather the opposite (Fig. S5 D).
Finally, we tested for asynchrony in poleward anaphase movements due to a reduction in poleward microtubule flux (Matos et al., 2009). To quantify the synchronicity of anaphase movements (Matos et al., 2009; Pereira and Maiato, 2010; Vukušić et al., 2019), we tracked kinetochores in RPE1 eGFP-CENP-A/eGFP-centrin-1 cells and quantified speeds of anaphase A (movements toward the poles) and B (movements of the poles relative to each other) and the spread of the chromosomes during their movements toward the poles. While KATNB1 depletion reduced anaphase A speeds and increased anaphase B speeds when compared with control-depleted cells, WDR62 loss did not change either of them (Fig. 6, E and F). In contrast, both WDR62 and KATNB1 depletion led to a wider spread of the kinetochore half-plates in their movements toward the spindle pole, indicating that both proteins are important for the synchronicity of poleward anaphase movements (Fig. 6 G, Video 7, and Video 8). Our results therefore validate the hypothesis that minus-end depolymerization required for flux plays an important role in equalizing forces in the spindle to ensure synchronous anaphase movements.
Video 7.
Sister-kinetochore oscillations of siCTRL-treated hTert-RPE1 GFP-centrin1/GFP-CENPA cells, showing tight metaphase plate and synchronous anaphase onset. Related to Fig. 6. Sampling rate = 15 s; scale bars = 5 µm. Play rate = 4 frames per second.
Video 8.
Sister-kinetochore oscillations of siWDR62-treated hTert-RPE1 GFP-centrin1/GFP-CENPA cells, showing wide metaphase plate and asynchronous anaphase onset with lagging chromosomes. Related to Fig. 6. Sampling rate = 15 s; scale bars = 5 µm. Play rate = 4 frames per second.
Discussion
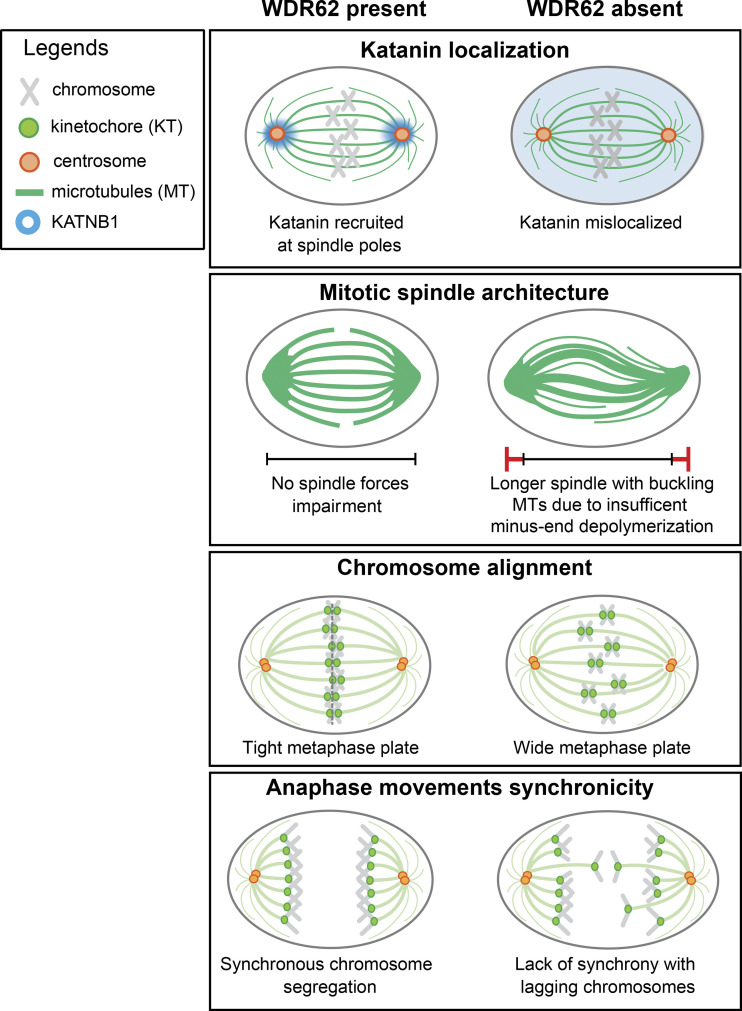
Most genes associated with primary microcephaly have a general impact on cell division. The exact cellular function of WDR62/MCPH2 is, however, still unclear. Based on our data, we propose that WDR62 ensures the correct localization of the katanin complex at spindle poles. The correct localization of this microtubule-severing activity at spindle poles ensures dynamic microtubules, prevents an overstabilization of k-fibers, and allows for efficient poleward microtubule flux rates. Finally, we demonstrate that loss of WDR62 or katanin impairs the synchronicity of anaphase poleward movements resulting in lagging chromosomes (see model, Fig. 7).
Figure 7.
WDR62 localizes KATNB1 at spindle poles to ensure synchronous anaphase. Model of how WDR62 ensures efficient minus-end depolymerization by localizing katanin at spindle poles to ensure tight metaphase plates and synchronous anaphase movements.
Our data indicate that WDR62 is localized at spindle poles, at or in the immediate vicinity of microtubule minus-ends, where it plays an essential role in the recruitment of katanin. This microtubule-severing complex fails to localize to spindle poles in both HeLa and RPE1 cells when WDR62 is depleted or lost, and our complementation assay indicates that katanin mislocalization is not caused by an off-target effect. Immunoblotting confirms that WDR62 controls only katanin localization, not the overall protein abundance. We propose that a deregulation of katanin is a major pathway causing primary microcephaly in humans. Indeed, this pathology can be caused by mutations in the KATNB1 gene itself (Mishra-Gorur et al., 2014) or by the loss of two microcephaly genes required for the enrichment of katanin at spindle poles: ASPM (Jiang et al., 2017) and WDR62 (this study). Our conclusions are reinforced by a parallel study demonstrating that recombinant WDR62 can recruit katanin to sever microtubules in vitro (Huang et al., 2021). Our dependence experiments indicate that WDR62 does not require ASPM to localize to spindle poles and that vice versa ASPM is unaffected by WDR62 depletion, indicating that they recruit katanin via separate pathways. A previous study based on mouse embryonic fibroblasts reported that ASPM and WDR62 form a complex at the mother centriole and play a crucial role in centrosome duplication (Jayaraman et al., 2016). While our data do not exclude a direct interaction between ASPM and WDR62, we found no evidence for strong centrosome duplication defects after WDR62 loss or depletion, nor was WDR62 enriched at centrioles. Rather, we found that WDR62 preferentially localizes to spindle microtubule parts that terminate at spindle poles, possibly at microtubule minus-ends. This is consistent with previous studies reporting that WDR62 is found at microtubule nucleation sites associated with centrosomes or not (Bogoyevitch et al., 2012).
At the functional level we demonstrate that WDR62 loss differentially affects spindle microtubules: it increased k-fiber stability while reducing the density of astral microtubules. Since WDR62 itself does not localize to astral microtubules, we speculate that this latter effect is indirect. One possibility could be that the increased incorporation of tubulin dimers into k-fibers diminishes the pool of free tubulin dimers, decreasing tubulin incorporation in astral microtubules. Our results are also consistent with a loss of katanin at spindle poles, as full katanin depletion results in an equivalent yet stronger pattern. These results, however, diverge from a previous study reporting that WDR62−/− mouse embryonic fibroblasts show a mild decrease in microtubule stability (Chen et al., 2014). The same study reported a mitotic arrest and unattached kinetochores in WDR62−/−, while our WDR62-depleted RPE1 or WDR62-deleted HeLa cells showed only a mild delay in mitotic progression, raising the possibility that human and murine cells might react differently to the loss of WDR62. One remarkable aspect is that WDR62 exclusively affected microtubule minus-end dynamics, reducing the rate of poleward microtubule flux, while leaving the k-fiber plus-end dynamics unaffected. We conclude that the two extremities of k-fibers can be regulated independently and are not necessarily mechanically coupled. Second, the results indicate that k-fiber stability is also determined by microtubule minus-end turnover, highlighting the emerging importance of minus-end binding proteins (Akhmanova and Steinmetz, 2019; Jiang et al., 2017; 2014) and attenuating the prevalent view that k-fiber dynamics is primarily controlled by microtubule plus-end dynamics (Akhmanova and Steinmetz, 2015).
How does the decrease in minus-end turnover in cells lacking WDR62 affect the function of the mitotic spindle? First, we find an increase in spindle length and buckling microtubules. Buckling is most likely the consequence of compressive forces that arise in the spindles when lengthening spindle microtubules are not severed (Tolić et al., 2019). Consistently, we also find buckling microtubules in cells depleted of KATNB1. In contrast to previous studies (Bogoyevitch et al., 2012; Miyamoto et al., 2017), but consistent with others (Chen et al., 2014; Jayaraman et al., 2016), we find no spindle orientation defects in cells lacking WDR62, in line with mouse experiments in which spindle orientation defects are not a major driver of primary microcephaly (Insolera et al., 2014). We also note that the increase in the average angle after WDR62 loss was either small (2°; Miyamoto et al., 2017) or based on only 12–14 cells in two independent experiments (Bogoyevitch et al., 2012), vs. >300 cells in five independent experiments in two different cell lines in the present study. We conclude that WDR62 depletion does not cause a major spindle orientation phenotype in a 2D tissue culture. This is surprising, given the reduction in astral microtubule density, which is critical for spindle orientation control. It is, however, possible that the increased spindle length seen in WDR62- or katanin-depleted cells compensates for the reduction in astral microtubules, allowing a sufficient interaction between cell cortex and astral microtubules. The consequences might be different in early murine neuronal stem cells that express low levels of the spindle-associated aurora-A activator TPX2 (Kufer et al., 2002), which controls the balance between spindle and astral microtubules (Vargas-Hurtado et al., 2019).
Nevertheless, the most striking phenotype we observe is that WDR62 loss leads to wider metaphase plates and asynchronous poleward anaphase movements. The increased kinetochore spread in anaphase results in lagging chromosomes that are integrated very late into the daughter nuclei. The monastrol-release experiments imply that these lagging chromosomes are not the results of an inefficient correction of erroneous kinetochore–microtubule attachments due to hyperstable k-fibers (Gregan et al., 2011; Lampson and Grishchuk, 2017). This suggests that inefficient error correction might correlate more with changes in microtubule plus-end turnover than with overall k-fiber stability (Bakhoum et al., 2009a), consistent with the observation that k-fiber stability and plus-end turnover are not necessarily correlated (Amaro et al., 2010). We postulate that in WDR62-depleted cells, asynchronous anaphase movements and lagging chromosomes are a direct consequence of reduced poleward microtubule flux rates. The decrease in flux rate is most likely due to the inability of WDR62-depleted cells to efficiently sever microtubules at spindle poles, resulting in a jam that slows down the motor-driven flux machinery (Steblyanko et al., 2020) and leads to buckling microtubules. Our data suggest that lagging chromosomes are caused by the reduction in flux rates, and that microtubule buckling is separate symptom of the same condition. A link between efficient microtubule flux rate and chromosome segregation had been predicted by mathematical models, which postulated that poleward flux equalizes the pulling forces within the mitotic spindles (Matos et al., 2009). Consistent with a role for flux in force equalization, we find that WDR62 depletion does not change the anaphase A or B speeds, but specifically increases the spatial spread of kinetochores in metaphase and during anaphase. We postulate that a main function of WDR62 is to localize katanin on the spindle poles to ensure synchronous anaphase movements via poleward microtubule flux.
Interestingly, depletion of katanin itself led to a very similar but not equivalent phenotype. In metaphase, katanin depletion led to the same reduction in poleward microtubule flux, microtubule buckling, and even a stronger microtubule stabilization, indicating that unlike Drosophila katanin (Zhang et al., 2007), human katanin actively regulates microtubule dynamics at spindle poles before anaphase onset. While katanin depletion also increased metaphase plate width at anaphase onset (Fig. 5, G and H), these effects were weaker than in WDR62-depleted cells and not significant within our sample size. This could be due to the effects of other katanin-pools, as it can also localize to microtubule plus-ends (Zhang et al., 2007), or to even higher suppression of spindle microtubule dynamics, which might limit the amplitude of chromosome movements (Fig. S2 D). During anaphase, katanin depletion led to not only a higher spread of chromosome, but also lower anaphase A and higher anaphase B speeds, an effect not seen after WDR62 depletion. This points to an additional role beyond the pool at spindle poles, consistent with the observation that Drosophila katanin is required for the “Pac-man”-like microtubule plus-end depolymerization during anaphase A (Zhang et al., 2007).
Based on our results, we postulate that WDR62 leads to defective chromosome segregation. Previous work had established that gain or loss of entire chromosomes (aneuploidy) in neural precursor cells can lead to microcephaly via premature differentiation in D. melanogaster (Gogendeau et al., 2015) or apoptosis in mice (Marthiens et al., 2013). Here, we speculate that WDR62 loss might lead to microcephaly not via gain or loss of entire chromosomes, but because of a late incorporation into the nucleus. Such events might lead to DNA damage due to incomplete nuclear envelope formation (de Castro et al., 2017) or to incorrect chromosomal positioning in the future interphasic daughter nuclei (Gerlich et al., 2003). Indeed, chromosome misalignment in metaphase and a high spread in anaphase has been linked to developmental defects in mice, suggesting that lagging chromosomes may similarly disrupt proper development in WDR62 mutants (Fonseca et al., 2019; Orr and Maiato, 2019). Since WDR62 also affects cilia length (Shohayeb et al., 2020; Zhang et al., 2019), this might lead to cumulative defects affecting correct brain development, without necessarily a severe disruption of chromosome segregation.
Materials and methods
Cell culture and drug treatments
HeLa Kyoto, HeLa H2B-GFP (Meraldi et al., 2004), HeLa H2B-GFP WDR62−/−, HeLa WDR62−/− (kind gift of K. Jiang, Wuhan University, China), hTert-RPE1, hTert-RPE1 EB3-GFP/H2B-mCherry (kind gift of W. Krek, ETH Zürich, Switzerland), hTert-RPE1 GFP-centrin1/GFP-CENPA (kind gift from A. Khodjakov, State University of New York) cell lines were cultured in DMEM (Thermo Fisher Scientific), supplemented with 10% FCS (Labforce), and 100 U/ml penicillin and 100 mg/ml streptomycin (Thermo Fisher Scientific). hTert-RPE1 PA-GFP-α-tubulin cells (Toso et al., 2009) and hTert-RPE1 stably transfected with pIC113-eGFP or pIC113-WDR62-eGFP plasmids encoding for eGFP or WDR62-eGFP were in addition supplemented with 600 µg/ml G418 (InvivoGen). To create HeLa H2B-GFP WDR62−/− CRISPR knockout cells, HeLa H2B-GFP cells were transiently transfected with X-tremeGENE 9 (Roche), and a plasmid encoding both the Cas9-mRFP enzyme and a gRNA targeting the first exon of the human WDR62 gene (target sequence: 5′-GCTATGCGCGGAACGATGCAGG-3′; Sigma-Aldrich). Single Cas9-mRFP–positive cells were selected by FACS 24 h after transfection and sorted into a 96-well dish. Growing colonies were tested by PCR for small deletions, sequenced to confirm WDR62 gene frameshift, and validated for loss of WDR62 expression by immunofluorescence. All cells used were female and were cultured at 37°C with 5% CO2 for maximum 6 wk in a humidified incubator. For live-cell imaging, cells were cultured in eight-, four-, or two-well Ibidi chambers (Vitaris) in Leibovitz’s L-15 medium (Thermo Fisher Scientific) supplemented with 10% FCS before imaging. The following drugs were added to the culturing or imaging medium: 10 µM MG132 for 30 min, 100 µM monastrol for 12–16 h, 200 ng/ml nocodazole for 15 min, 8 nM taxol (all Sigma-Aldrich) for 12 h, and 50 nM SiR-tubulin (Spirochrome) for 4 h.
siRNA and plasmid transfections
For protein depletions, cells were transfected for 48 h (72 h in case of siCAPD2) with 20 nM siRNAs using Opti-MEM and Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. The medium was replaced 24 h after transfection. The following sense strands of validated siRNA duplexes were used (all Qiagen unless indicated): siASPM (validation in Fig. S2), 5′-GCUAUAUGUCAGCGUACUATT-3′; siCAPD2 (Hirota et al., 2004), 5′-CCAUAUGCUCAGUGCUACATT-3′; siKATNB1 (validation in Fig. S2), 5′-GCCUGGAUUUCCACCCGUATT-3′; siWDR62 (Dharmacon; validation in Fig. S1), 5′-AGACAAAGGUGACGAGCAC-3′; and as a negative control, either siCTRL (AllStars Negative Control siRNA; Qiagen), Proprietary or siLaminA (Qiagen), 5′GAAGGAGGGUGACCUGAUA-3′. For transient exogenous gene expression, cells were transfected using X-tremeGENE 9 DNA Transfection Reagent (Roche) with a ratio of 3:1 µl/µg plasmid of DNA and analyzed 30 h after transfection. The pIC113-WDR62-eGFP plasmid encoding for an eGFP-tagged WDR62 was synthesized and subcloned by GeneArt (Thermo Fisher Scientific).
Monitoring of chromosome segregation by live-cell imaging
To monitor mitotic progression and chromosome segregation, HeLa H2B-eGFP, HeLa H2B-eGFP WDR62−/−, or hTert-RPE1 EB3-GFP/H2B-mCherry cells treated with siCTRL or siWDR62 were seeded into Ibidi chambers (Vitaris) supplemented with imaging medium 2–3 h before imaging and recorded on a Nikon Eclipse Ti-E wide-field microscope (Nikon) equipped with a GFP/mCherry filter set (Chroma Technology Corp.), an Orca Flash 4.0 complementary metal-oxide-semiconductor camera (Hamamatsu), and an environmental chamber using NIS software (Nikon). Cells were imaged in the GFP and mCherry channel for 9 h at 37°C, at 1-min intervals, in 9 steps of 2 µm Z-stacks, using a 60× (NA 1.51) oil objective and 2 × 2 binning. Alternatively, cells were released from monastrol arrest and imaged under the same conditions for 1 h and 15 min. Time-lapse movies were analyzed manually with NIS Elements software to quantify mitotic timing and segregation errors.
Spindle microtubule depolymerization assay
To quantify spindle microtubule stability, live hTert-RPE1 cells were seeded in Ibidi eight-well chamber (Vitaris) and depleted of the indicated protein. Before imaging, 50 nM SiR-tubulin (Spirochrome) was added for 4 h to label microtubules, and 10 µM MG132 was added for 30 min to prevent mitotic exit. Imaging medium containing 200 ng/ml nocodazole, 50 nM SiR-tubulin, and 10 µM MG132 was added upon starting time-lapse imaging. Cells were imaged every 1 min for 15 min using a 100× (NA 1.4) oil objective on an Olympus DeltaVision wide-field microscope (GE Healthcare) equipped with an environmental chamber maintained at 37°C and with DAPI/FITC/TRITC/Cy5 filter set (Chroma Technology Corp.) and Coolsnap HQ2 charge-coupled device (CCD) camera (Roper Scientific) running Softworx (GE Healthcare). Z-stacks of 15-µm thickness with z-slices separated by 0.5 µm were imaged in the Cy5 channel with 2 × 2 binning to track for microtubule depolymerization. 4D images (XYZT) obtained were deconvolved using Softworx (GE Healthcare) in conservative mode, and the intensity decay of the whole spindle was quantified using an automated ImageJ plugin (coded in NetBeans IDE, Apache Software Foundation; available at https://github.com/AmandaGuerreiro/WDR62_2020). The plugin segments each cell based on Otsu’s method and performs a sum intensity projection along the z-axis for all time points. Using RStudio, the values of 4D image pixels were summed, resulting in a fluorescence intensity signal over time. Microtubule loss was expressed as a percentage of the decay of the SiR-tubulin signal normalized to the first time point (100%). Statistical analysis of microtubule depolymerization was done using repeated paired t test and a 95% CI over time.
Kinetochore tracking (KT) assay
hTert-RPE1 GFP-centrin1/GFP-CENPA were treated with the indicated siRNA, and metaphase cells were imaged for 15 min every 15 s using a 100× (NA 1.4) oil objective on an Olympus DeltaVision wide-field microscope (GE Healthcare) equipped with an environmental chamber maintained at 37°C and a GFP/RFP filter set (Chroma Technology Corp.) and Coolsnap HQ2 CCD camera (Roper Scientific) running Softworx (GE Healthcare). Z-stacks of 15-µm thickness with z-slices separated by 0.5 µm were imaged in the GFP channel with 2 × 2 binning to track for kinetochores from metaphase until anaphase. 4D images (XYZT) obtained were deconvolved in conservative mode and cropped using Softworx (GE Healthcare).
For KT in metaphase, 4D images (XYZT) were analyzed using an automated KT code written in MatLab 2013b (MathWorks; Jaqaman et al., 2010). The latest code is available under https://github.com/cmcb-warwick, and the parameters described in Olziersky et al. (2018). Briefly, the output of this analysis is the frame-to-frame displacement of sister-kinetochores and their relative distance from the center of the metaphase plate. We extracted the interkinetochore distance of sister-kinetochore pairs and used an autocorrelation function to quantify the regularity of the sister-kinetochore oscillations along the spindle axis. Kinetochore velocities were calculated by plotting the distribution of all sister-kinetochore displacements and calculating the SD of this distribution.
For KT in anaphase, 4D acquisitions (XYZT) were processed using Imaris 9.5.1 (Bitplane) to segment kinetochores and centrioles during metaphase and anaphase. All the positions through time were analyzed using a custom-made framework written in Matlab 2019b (available under https://github.com/AmandaGuerreiro/WDR62_2020). Anaphase onset was automatically detected by finding the abrupt slope change of the pole-to-pole distance during the time course. Accuracy detection was assessed by the user and manually shifted if necessary. Anaphase A speed for each pole was defined as the slope of a linear fit of the kinetochore-to-pole mean distance during 120 s after onset. Anaphase B speed was defined as the slope of the pole-to-pole distance during the same time after onset. Kinetochore spread was quantified by orthogonal projection of all the kinetochores along the pole-to-pole axis and was represented as the maximum distance between the kinetochores over time in metaphase (Fig. 5 G), the average (obtained from the SD) width in the last 30 s before anaphase onset (Fig. 5 H), and the maximum distance between kinetochores during anaphase (Fig. 6 G).
Poleward microtubule flux measurement
hTert-RPE1 PA-GFP-α-tubulin cells were incubated with 50 nM SiR-DNA in the last 2 h and 10 µM MG132 in the last 30 min. Cells incubated were imaged for ≤2 h. Single focal planes of 150-nm pixel size were acquired using a 60× (NA 1.4) CFI Plan Apochromat oil objective on a Nikon A1r point scanning confocal microscope equipped with a 37°C heating chamber and running NIS elements software. Half-spindles were photoactivated with a 500-ms, 405-nm laser pulse at 30–80% intensity depending on the PA-GFP-α-tubulin expression levels, using a 1-pixel-thick and 100-pixel-long region of interest stretched across the spindle. Single focal planes were imaged every 20 s for 4 min. Photoactivated kinetochore-microtubule bundles were tracked for 80 s. By computing manually the mean distance between the photoactivation mark on kinetochore-microtubule bundles and the corresponding spindle poles at different time points, we calculated the mean displacement of the photoactivation mark over time. Note that since spindle length did not change during the recording period, the rate of progression toward the spindle pole and the rate of displacement away from the metaphase plate is equivalent.
Immunofluorescence
For the cold-stable assay, hTert-RPE1 and HeLa cells were treated with ice-cold medium for 6 or 20 min. To label microtubules, cells were rinsed with cytoskeleton buffer (10 mM MES, 150 mM NaCl2, 5 mM MgCl2, and 5 mM glucose; Sigma-Aldrich) before and 2 × 10 min after fixation with a solution composed of 3% formaldehyde, 0.05% glutaraldehyde, and 0.1% Triton X-100 (Sigma-Aldrich) for 15 min at 4°C. The same fixation method was also used to stain astral microtubules. To induce acentrosomal spindles, HeLa cells were treated for 12 h with 8 nM taxol or DMSO. For minus-end quantifications, cells were fixed with −20°C methanol for 7 min and rinsed once for 5 min with 0.15% Triton X-100 in PBS. For MCAK and CAPD2 quantifications, cells were fixed with 10% formaldehyde, 25 M of 8% PIPES, 0.25 M of 4% EGTA, and 2% Triton X-100 (Sigma-Aldrich) in 76% ddH2O for 15 min at RT; subsequent extraction was performed in blocking buffer supplemented with 0.5% Triton X-100 for 2 × 10 min followed by PBS with 0.1% Triton X-100 for 3 × 5 min. After fixation, cells were blocked overnight at 4°C in PBS + 3% BSA and 1% N3Na (Applichem). Subsequent washings were performed in PBS with 0.05% Tween-20 for 3 × 10 min and in blocking buffer supplemented with 0.1% Triton X-100 for 3 × 10 min for MCAK and CAPD2 quantifications. Labeling steps were performed in blocking solution for 1 h at RT for primary antibodies and 30 min at RT for secondary antibodies. Coverslips were mounted on slides with DAPI Vectashield mounting medium (Vector Laboratories). The following primary antibodies were used: mouse anti-α-tubulin (1:500; Sigma-Aldrich), recombinant human anti-α-tubulin (1 µg/ml; Guerreiro and Meraldi, 2019), rabbit anti-γ-tubulin (1:2,000; Wilhelm et al., 2019), mouse anti-γ-tubulin (1:1,000; Sigma-Aldrich), rabbit anti-ASPM (1:100; Novus Biologicals), guinea pig anti-CENP-C (1:1,000; MBL), mouse anti-centrin-1 (1:2,000; Merck Millipore), rabbit anti-KATNB1 (1:250; Proteintech), rabbit anti-CAPD2 (1:100; Abcam), rabbit anti-NuMA (1:100; Abcam), mouse anti-pericentrin (1:1,000; Abcam), and rabbit anti-WDR62 (1:1,000; Bethyl). For secondary antibodies, cross-adsorbed Alexa Fluor–conjugated antibodies (1:400; Invitrogen) were used. Pictures were acquired using 60× and 100× (NA 1.4) oil objectives on an Olympus DeltaVision wide-field microscope (GE Healthcare) equipped with a DAPI/FITC/TRITC/Cy5 filter set (Chroma Technology Corp.) and Coolsnap HQ2 CCD camera (Roper Scientific) running Softworx (GE Healthcare). Z-stacks of 12.80-µm thickness were imaged with z-slices separated by 0.2 µm. 3D images stacks were deconvolved using Softworx (GE Healthcare) in conservative mode.
STED nanoscopy and confocal microscopy
Multicolor 2D-STED imaging was performed with a TCS SP8 STED 3× microscope (Leica) at 21°C using a STED motorized glycerol immersion objective (HC PL Apo 93×/NA 1.30 motCOR). Fluorescence-labeled samples were mounted in Prolong Antifade Gold (Thermo Fisher Scientific) between a coverslip (0.170 ± 0.01 mm thick; Hecht-Assistent) and a microscope slide and sealed with nail polish. Excitation was performed with a white light laser (WLL), and depletion with either a continuous 592-nm laser or a 775-nm pulsed laser. Excitation and depletion lasers were calibrated with the STED Expert Alignment Mode and Abberior gold nanoparticles (80 nm in diameter) before starting each imaging session, or with the STED Auto Beam Alignment tool during imaging sessions (Leica LAS X software). Multicolor 2D-STED imaging was made sequentially using excitation at 638 nm (WLL) and a STED 775 depletion laser line for Abberior STAR RED anti-rabbit antibody, followed by excitation at 580 nm (WLL) and a STED 775 depletion laser line for Abberior STAR 580 anti-mouse antibody, and then by excitation at 488 nm (WLL) and a STED 592 depletion laser line for Alexa Fluor 488 anti-human antibody. Detection signals were collected between 647 and 677 nm for STAR RED, between 591 and 612 nm for STAR 580, and between 492 and 562 nm for AF488 using highly sensitive Leica Hybrid Detectors with a fixed gain and offset (100 mV and 0, respectively). Time-gated detection was used for all fluorophores (0.50–6.00 ns). Acquisitions were performed with a line average of 4, a speed of 400 Hz, and an optimized pixel size. 2D-STED images were deconvolved using the Leica Lightning Mode (LAS X software) and analyzed with ImageJ/Fiji (National Institutes of Health). Z-stack confocal acquisitions were obtained in the same conditions, but without depletion and time-gated detection, with an optimized pinhole aperture and by respecting Nyquist criterion.
Image processing and analysis
Immunofluorescence images from the cold-stable assay were analyzed blindly: the acquired images were visualized in 3D using Imaris (Bitplane), and cells were categorized into three different classes according to the abundance of kinetochore fibers. For astral microtubules and minus-end immunofluorescence quantifications in metaphase, 3D images were analyzed with ImageJ/Fiji using a plugin developed with NetBeans IDE (v.8.2). Briefly, a mask was created on centrosomes using yen dark thresholding on sum projections. Quantification of fluorescence was performed for each channel using the mask. For kinetochore immunofluorescence quantification in interphase, 3D images were analyzed manually with ImageJ/Fiji using yen dark thresholding on sum projections. For spindle orientation quantification in immunofluorescence, 3D images were analyzed with ImageJ/Fiji. A plugin was used to calculate the angle from two points in regions of interest defined in the Z-stack at centrosome position.
Immunoblotting
For cell extracts, cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.3, 150 mM NaCl, 0.1 mM EDTA, 1% Triton X-100, and 0.5 mM PMSF) supplemented with Protease Inhibitor Cocktail and PhosSTOP phosphatase inhibitor (Sigma-Aldrich). Cell lysates were incubated on ice (30 min) and cleared by centrifugation (16,000 g, 10 min at 4°C). Protein concentrations were determined by Bradford assay (Thermo Fisher Scientific). 2× Laemmli buffer was added before SDS-PAGE and not boiled. Proteins were separated on 4–15% gradient TGX precast gels (Bio-Rad), transferred onto nitrocellulose membrane by wet blotting, and blocked for 1 h at RT with 5% nonfat milk powder in PBS. Subsequent washings were performed in PBS with 0.05% Tween-20 for 3 × 10 min. The following primary antibodies were used: rabbit anti-KATNB1 (1:500; Proteintech), rabbit anti-WDR62 (1:500; Bethyl), and mouse anti-β-actin (1:1,000; Merck Millipore). The following HRP-conjugated secondary antibodies were used: ECL anti-rabbit IgG and anti-mouse IgG (Thermo Fisher Scientific). Protein bands were detected using the ECL Prime Western Blotting Detection reagent (GE Healthcare) and a PXi/PXi Touch luminescence detector (Syngene). Protein bands were quantified with ImageJ as described in the method outlined at https://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/. Specific protein signals were normalized to β-actin.
Statistical analysis
Data were quantified using ImageJ/Fiji, Imaris software (v7.7 or v9.3), Matlab (versions 2013b, 2018a, or 2019b), and RStudio (v1.2). Statistical analysis was performed in Prism (GraphPad, v8.4), Matlab (KT assay), or RStudio (kinetochore microtubule depolymerization assay and frequency distribution of spindle orientation with pole-to-pole distance). Frequency distributions in RStudio were computed using kernel density estimate function.
Online supplemental material
Fig. S1 shows WDR62 deletion and depletion efficiency. Fig. S2 shows that KATNB1 depletion mimics WDR62 depletion. Fig. S3 shows that WDR62−/− HeLa cells show no difference in spindle orientation or pole-to-pole distance, and spindle length of MG132-arrested RPE1 cells are constant. Fig. S4 shows validation of CAPD2 depletion in RPE1 cells. Fig. S5 shows that loss of WDR62 in HeLa cells does not affect mitotic timing but increases the percentage of lagging chromosomes. Video 1, Video 2, and Video 3 show microtubule poleward flux after photoactivation of hTert-RPE1 PA-GFP-α-tubulin cells treated with siCTRL, siKATNB1, and siWDR62, respectively. Video 4 shows no error of chromosome segregation in hTert-RPE1 EB3-GFP/H2B-mCherry cells treated with siCTRL. Video 5 shows lagging chromosomes in anaphase in hTert-RPE1 EB3-GFP/H2B-mCherry cells treated with siWDR62. Video 6 shows a DNA bridge in anaphase in hTert-RPE1 EB3-GFP/H2B-mCherry cells treated with siWDR62. Video 7 shows sister-kinetochore oscillations of siCTRL treated hTert-RPE1 GFP-centrin/GFP-CENPA cells, showing tight metaphase plate and synchronous anaphase onset. Video 8 shows sister-kinetochore oscillations of siWDR62 treated hTert-RPE1 GFP-centrin1/GFP-CENPA cells, showing wide metaphase plate and asynchronous anaphase onset with lagging chromosomes.
Supplementary Material
Acknowledgments
We thank A. Khodjakov (State University of New York) and the late W. Krek (ETH Zürich, Switzerland) for cell lines; K. Jiang (Wuhan University, China) for sharing results before publication and the HeLa WDR62−/− cells; the Bioimaging Facility at the Medical Faculty, University of Geneva, Switzerland, for microscopy support; M. Hoveyda (University of Geneva) for initiating the investigation of WDR62 and spindle stability; L. Brodier (University of Geneva) for technical discussions and help with R and ImageJ plugins; D. Dudka (University of Pennsylvania), M. Gotta (University of Geneva), and T. Wilhelm (Curie Institute, France) for critical discussions; and members of the Meraldi laboratory for support and technical help.
Work in the Meraldi laboratory of A. Guerreiro, F. De Sousa, D. Ivanova, A. Eskat, and P. Meraldi is supported by a Swiss National Science Foundation project grant (31003A_179413) and the University of Geneva.
The authors declare no competing financial interests.
Author contributions: The project was initiated by A. Guerreiro and P. Meraldi and directed by P. Meraldi; A. Guerreiro performed all experiments; F. De Sousa contributed to chromosome segregation experiments and immunoblotting; N. Liaudet wrote the anaphase tracking code; A. Eskat generated the HeLa H2B-GFP WDR62−/− cell line; D. Ivanova contributed to spindle size and chromosome segregation experiments; A. Guerreiro, F. De Sousa, and P. Meraldi analyzed and interpreted all the results with contribution from N. Liaudet; and A. Guerreiro and P. Meraldi wrote the manuscript.
References
- Akhmanova, A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Akhmanova, A., and Steinmetz M.O.. 2019. Microtubule minus-end regulation at a glance. J. Cell Sci. 132:jcs227850. 10.1242/jcs.227850 [DOI] [PubMed] [Google Scholar]
- Amaro, A.C., Samora C.P., Holtackers R., Wang E., Kingston I.J., Alonso M., Lampson M., McAinsh A.D., and Meraldi P.. 2010. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12:319–329. 10.1038/ncb2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum, S.F., Genovese G., and Compton D.A.. 2009a. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 19:1937–1942. 10.1016/j.cub.2009.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum, S.F., Thompson S.L., Manning A.L., and Compton D.A.. 2009b. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11:27–35. 10.1038/ncb1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto, R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., and Raff J.W.. 2006. Flies without centrioles. Cell. 125:1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bilgüvar, K., Oztürk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A.O., Gökben S., et al. 2010. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 467:207–210. 10.1038/nature09327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch, M.A., Yeap Y.Y.C., Qu Z., Ngoei K.R., Yip Y.Y., Zhao T.T., Heng J.I., and Ng D.C.H.. 2012. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J. Cell Sci. 125:5096–5109. 10.1242/jcs.107326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, J., Roberts E., Mochida G.H., Hampshire D.J., Scott S., Askham J.M., Springell K., Mahadevan M., Crow Y.J., Markham A.F., et al. 2002. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32:316–320. 10.1038/ng995 [DOI] [PubMed] [Google Scholar]
- Bond, J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355. 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Bond, J., Scott S., Hampshire D.J., Springell K., Corry P., Abramowicz M.J., Mochida G.H., Hennekam R.C.M., Maher E.R., Fryns J.-P., et al. 2003. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am. J. Hum. Genet. 73:1170–1177. 10.1086/379085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster, D.W., Zhang D., and Sharp D.J.. 2007. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol. Biol. Cell. 18:3094–3104. 10.1091/mbc.e06-11-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.-F., Zhang Y., Wilde J., Hansen K.C., Lai F., and Niswander L.. 2014. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5:3885. 10.1038/ncomms4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Wainman A., and Raff J.W.. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16:611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- David, A.F., Roudot P., Legant W.R., Betzig E., Danuser G., and Gerlich D.W.. 2019. Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth. J. Cell Biol. 218:2150–2168. 10.1083/jcb.201805044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro, I.J., Gil R.S., Ligammari L., Di Giacinto M.L., and Vagnarelli P.. 2017. CDK1 and PLK1 coordinate the disassembly and reassembly of the nuclear envelope in vertebrate mitosis. Oncotarget. 9:7763–7773. 10.18632/oncotarget.23666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudka, D., Castrogiovanni C., Liaudet N., Vassal H., and Meraldi P.. 2019. Spindle-Length-Dependent HURP Localization Allows Centrosomes to Control Kinetochore-Fiber Plus-End Dynamics. Curr. Biol. 29:3563–3578.e6. 10.1016/j.cub.2019.08.061 [DOI] [PubMed] [Google Scholar]
- Elting, M.W., Hueschen C.L., Udy D.B., and Dumont S.. 2014. Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol. 206:245–256. 10.1083/jcb.201401091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, C.L., Malaby H.L.H., Sepaniac L.A., Martin W., Byers C., Czechanski A., Messinger D., Tang M., Ohi R., Reinholdt L.G., and Stumpff J.. 2019. Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. J. Cell Biol. 218:1148–1163. 10.1083/jcb.201807228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., Upton K., and Compton D.A.. 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15:1827–1832. 10.1016/j.cub.2005.08.065 [DOI] [PubMed] [Google Scholar]
- Gerlich, D., Beaudouin J., Kalbfuss B., Daigle N., Eils R., and Ellenberg J.. 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 112:751–764. 10.1016/S0092-8674(03)00189-2 [DOI] [PubMed] [Google Scholar]
- Godek, K.M., Kabeche L., and Compton D.A.. 2015. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat. Rev. Mol. Cell Biol. 16:57–64. 10.1038/nrm3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogendeau, D., Siudeja K., Gambarotto D., Pennetier C., Bardin A.J., and Basto R.. 2015. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 6:8894. 10.1038/ncomms9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, M.B., Howard L., and Compton D.A.. 2001. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 152:425–434. 10.1083/jcb.152.3.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan, J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., and Cimini D.. 2011. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 21:374–381. 10.1016/j.tcb.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, A., and Meraldi P.. 2019. AA344 and AA345 antibodies recognize the microtubule network in human cells by immunofluorescence. Antibody Rep. 2:e17. 10.24450/journals/abrep.2019.e17 [DOI] [Google Scholar]
- Hirota, T., Gerlich D., Koch B., Ellenberg J., and Peters J.-M.. 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117:6435–6445. 10.1242/jcs.01604 [DOI] [PubMed] [Google Scholar]
- Hu, D.J.-K., and Jasper H.. 2019. Control of Intestinal Cell Fate by Dynamic Mitotic Spindle Repositioning Influences Epithelial Homeostasis and Longevity. Cell Rep. 28:2807–2823.e5. 10.1016/j.celrep.2019.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W.F., Pomp O., Ben-Omran T., Kodani A., Henke K., Mochida G.H., Yu T.W., Woodworth M.B., Bonnard C., Raj G.S., et al. 2014. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 84:1240–1257. 10.1016/j.neuron.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Liang Z., Guan C., Hua S., and Jiang K.. 2021. WDR62 regulates spindle dynamics as an adaptor protein between TPX2/Aurora A and katanin. J. Cell Biol. In press. 10.1083/jcb.202007167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, M.S., Baig S.M., Neumann S., Peche V.S., Szczepanski S., Nürnberg G., Tariq M., Jameel M., Khan T.N., Fatima A., et al. 2013. CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Hum. Mol. Genet. 22:5199–5214. 10.1093/hmg/ddt374 [DOI] [PubMed] [Google Scholar]
- Insolera, R., Bazzi H., Shao W., Anderson K.V., and Shi S.-H.. 2014. Cortical neurogenesis in the absence of centrioles. Nat. Neurosci. 17:1528–1535. 10.1038/nn.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman, K., King E.M., Amaro A.C., Winter J.R., Dorn J.F., Elliott H.L., McHedlishvili N., McClelland S.E., Porter I.M., Posch M., et al. 2010. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 188:665–679. 10.1083/jcb.200909005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, D., Bae B.-I., and Walsh C.A.. 2018. The Genetics of Primary Microcephaly. Annu. Rev. Genomics Hum. Genet. 19:177–200. 10.1146/annurev-genom-083117-021441 [DOI] [PubMed] [Google Scholar]
- Jayaraman, D., Kodani A., Gonzalez D.M., Mancias J.D., Mochida G.H., Vagnoni C., Johnson J., Krogan N., Harper J.W., Reiter J.F., et al. 2016. Microcephaly Proteins Wdr62 and Aspm Define a Mother Centriole Complex Regulating Centriole Biogenesis, Apical Complex, and Cell Fate. Neuron. 92:813–828. 10.1016/j.neuron.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F.M., Heck A.J.R., Hoogenraad C.C., and Akhmanova A.. 2014. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 28:295–309. 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Jiang, K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., Heck A.J.R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19:480–492. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P.2016. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology (Basel). 5:44. 10.3390/biology5040044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M.A., Toso R.J., Thrower D., and Wilson L.. 1993. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc. Natl. Acad. Sci. USA. 90:9552–9556. 10.1073/pnas.90.20.9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda, K., McAinsh A.D., and Cross R.A.. 2012. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol. Open. 1:12–18. 10.1242/bio.2011012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani, A., Yu T.W., Johnson J.R., Jayaraman D., Johnson T.L., Al-Gazali L., Sztriha L., Partlow J.N., Kim H., Krup A.L., et al. 2015. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 4:e07519. 10.7554/eLife.07519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer, T.A., Sillje H.H.W., Körner R., Gruss O.J., Meraldi P., and Nigg E.A.. 2002. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158:617–623. 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., and Grishchuk E.L.. 2017. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology (Basel). 6:1. 10.3390/biology6010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., Renduchitala K., Khodjakov A., and Kapoor T.M.. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lancaster, M.A., and Knoblich J.A.. 2012. Spindle orientation in mammalian cerebral cortical development. Curr. Opin. Neurobiol. 22:737–746. 10.1016/j.conb.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M.S., and Holland A.J.. 2018. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 32:620–638. 10.1101/gad.314351.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, N.R., Yeap Y.Y.C., Zhao T.T., Yip Y.Y., Wong S.C., Xu D., Ang C.-S., Williamson N.A., Xu Z., Bogoyevitch M.A., and Ng D.C.H.. 2015. Opposing roles for JNK and Aurora A in regulating the association of WDR62 with spindle microtubules. J. Cell Sci. 128:527–540. 10.1242/jcs.157537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, N.R., Yeap Y.Y.C., Ang C.-S., Williamson N.A., Bogoyevitch M.A., Quinn L.M., and Ng D.C.H.. 2016. Aurora A phosphorylation of WD40-repeat protein 62 in mitotic spindle regulation. Cell Cycle. 15:413–424. 10.1080/15384101.2015.1127472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius, G., Reymond L., D’Este E., Masharina A., Göttfert F., Ta H., Güther A., Fournier M., Rizzo S., Waldmann H., et al. 2014. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods. 11:731–733. 10.1038/nmeth.2972 [DOI] [PubMed] [Google Scholar]
- Marthiens, V., Rujano M.A., Pennetier C., Tessier S., Paul-Gilloteaux P., and Basto R.. 2013. Centrosome amplification causes microcephaly. Nat. Cell Biol. 15:731–740. 10.1038/ncb2746 [DOI] [PubMed] [Google Scholar]
- Matos, I., Pereira A.J., Lince-Faria M., Cameron L.A., Salmon E.D., and Maiato H.. 2009. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol. 186:11–26. 10.1083/jcb.200904153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHedlishvili, N., Wieser S., Holtackers R., Mouysset J., Belwal M., Amaro A.C., and Meraldi P.. 2012. Kinetochores accelerate centrosome separation to ensure faithful chromosome segregation. J. Cell Sci. 125:906–918. 10.1242/jcs.091967 [DOI] [PubMed] [Google Scholar]
- Meraldi, P.2016. Centrosomes in spindle organization and chromosome segregation: a mechanistic view. Chromosome Res. 24:19–34. 10.1007/s10577-015-9508-2 [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Draviam V.M., and Sorger P.K.. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60. [DOI] [PubMed] [Google Scholar]
- Mishra-Gorur, K., Çağlayan A.O., Schaffer A.E., Chabu C., Henegariu O., Vonhoff F., Akgümüş G.T., Nishimura S., Han W., Tu S., et al. 2014. Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron. 84:1226–1239. 10.1016/j.neuron.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J.1989. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 109:637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Akutsu S.N., Fukumitsu A., Morino H., Masatsuna Y., Hosoba K., Kawakami H., Yamamoto T., Shimizu K., Ohashi H., and Matsuura S.. 2017. PLK1-mediated phosphorylation of WDR62/MCPH2 ensures proper mitotic spindle orientation. Hum. Mol. Genet. 26:4429–4440. 10.1093/hmg/ddx330 [DOI] [PubMed] [Google Scholar]
- Nam, H.-J., and van Deursen J.M.. 2014. Cyclin B2 and p53 control proper timing of centrosome separation. Nat. Cell Biol. 16:538–549. 10.1038/ncb2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano, M., and Basto R.. 2017. Consequences of Centrosome Dysfunction During Brain Development. Adv. Exp. Med. Biol. 1002:19–45. 10.1007/978-3-319-57127-0_2 [DOI] [PubMed] [Google Scholar]
- Nicholas, A.K., Khurshid M., Désir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A., et al. 2010. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42:1010–1014. 10.1038/ng.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A., Čajánek L., and Arquint C.. 2014. The centrosome duplication cycle in health and disease. FEBS Lett. 588:2366–2372. 10.1016/j.febslet.2014.06.030 [DOI] [PubMed] [Google Scholar]
- Noatynska, A., Gotta M., and Meraldi P.. 2012. Mitotic spindle (DIS)orientation and DISease: cause or consequence? J. Cell Biol. 199:1025–1035. 10.1083/jcb.201209015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill, R.S., Schoborg T.A., and Rusan N.M.. 2018. Same but different: pleiotropy in centrosome-related microcephaly. Mol. Biol. Cell. 29:241–246. 10.1091/mbc.E17-03-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olziersky, A.-M., Smith C.A., Burroughs N., McAinsh A.D., and Meraldi P.. 2018. Mitotic live-cell imaging at different timescales. Methods Cell Biol. 145:1–27. 10.1016/bs.mcb.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Orr, B., and Maiato H.. 2019. No chromosome left behind: The importance of metaphase alignment for mitotic fidelity. J. Cell Biol. 218:1086–1088. 10.1083/jcb.201902041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, A.J., and Maiato H.. 2010. Improved kymography tools and its applications to mitosis. Methods. 51:214–219. 10.1016/j.ymeth.2010.01.016 [DOI] [PubMed] [Google Scholar]
- Prosser, S.L., and Pelletier L.. 2017. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18:187–201. 10.1038/nrm.2016.162 [DOI] [PubMed] [Google Scholar]
- Ramdas Nair, A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B., and Cabernard C.. 2016. The Microcephaly-Associated Protein Wdr62/CG7337 Is Required to Maintain Centrosome Asymmetry in Drosophila Neuroblasts. Cell Rep. 14:1100–1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Shohayeb, B., Ho U., Yeap Y.Y., Parton R.G., Millard S.S., Xu Z., Piper M., and Ng D.C.H.. 2020. The association of microcephaly protein WDR62 with CPAP/IFT88 is required for cilia formation and neocortical development. Hum. Mol. Genet. 29:248–263. 10.1093/hmg/ddz281 [DOI] [PubMed] [Google Scholar]
- Sikirzhytski, V., Magidson V., Steinman J.B., He J., Le Berre M., Tikhonenko I., Ault J.G., McEwen B.F., Chen J.K., Sui H., et al. 2014. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol. 206:231–243. 10.1083/jcb.201401090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth, W.T., Nardi I.K., Scholl L.M., and Cimini D.. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 4:e6564. 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth, W.T., Nardi I.K., Paul R., Mogilner A., and Cimini D.. 2012. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell. 23:401–411. 10.1091/mbc.e11-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir, J.-H., Pütz M., Daly O., Morrison C.G., Dunning M., Kilmartin J.V., and Gergely F.. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203:747–756. 10.1083/jcb.201309038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R.V., Skeen V.P., and Salmon E.D.. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steblyanko, Y., Rajendraprasad G., Osswald M., Eibes S., Jacome A., Geley S., Pereira A.J., Maiato H., and Barisic M.. 2020. Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J. 39:e105432. 10.15252/embj.2020105432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, S.L., and Compton D.A.. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672. 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolić, I.M., Novak M., and Pavin N.. 2019. Helical Twist and Rotational Forces in the Mitotic Spindle. Biomolecules. 9:132. 10.3390/biom9040132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso, A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., and McAinsh A.D.. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184:365–372. 10.1083/jcb.200809055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima, F., and Nishida E.. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26:1487–1498. 10.1038/sj.emboj.7601599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Hurtado, D., Brault J.-B., Piolot T., Leconte L., Da Silva N., Pennetier C., Baffet A., Marthiens V., and Basto R.. 2019. Differences in Mitotic Spindle Architecture in Mammalian Neural Stem Cells Influence Mitotic Accuracy during Brain Development. Curr. Biol. 29:2993–3005.e9. 10.1016/j.cub.2019.07.061 [DOI] [PubMed] [Google Scholar]
- Vukušić, K., Buđa R., and Tolić I.M.. 2019. Force-generating mechanisms of anaphase in human cells. J. Cell Sci. 132:jcs231985. 10.1242/jcs.231985 [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., Cai S., and Khodjakov A.. 2010. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol. 11:91–102. 10.1038/nrm2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, T., Olziersky A.-M., Harry D., De Sousa F., Vassal H., Eskat A., and Meraldi P.. 2019. Mild replication stress causes chromosome mis-segregation via premature centriole disengagement. Nat. Commun. 10:3585. 10.1038/s41467-019-11584-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, C.G., and Basto R.. 2014. Microcephaly. Curr. Biol. 24:R1109–R1111. 10.1016/j.cub.2014.09.063 [DOI] [PubMed] [Google Scholar]
- Xu, D., Zhang F., Wang Y., Sun Y., and Xu Z.. 2014. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Rep. 6:104–116. 10.1016/j.celrep.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Yu, T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topçu M., McDonald M.T., Barry B.J., Felie J.M., et al. 2010. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 42:1015–1020. 10.1038/ng.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., Rogers G.C., Buster D.W., and Sharp D.J.. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242. 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Yang S.-L., Yang M., Herrlinger S., Shao Q., Collar J.L., Fierro E., Shi Y., Liu A., Lu H., et al. 2019. Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 10:2612–2614. 10.1038/s41467-019-10497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.