Figure 3. RAD51 enhances interhomolog repair.
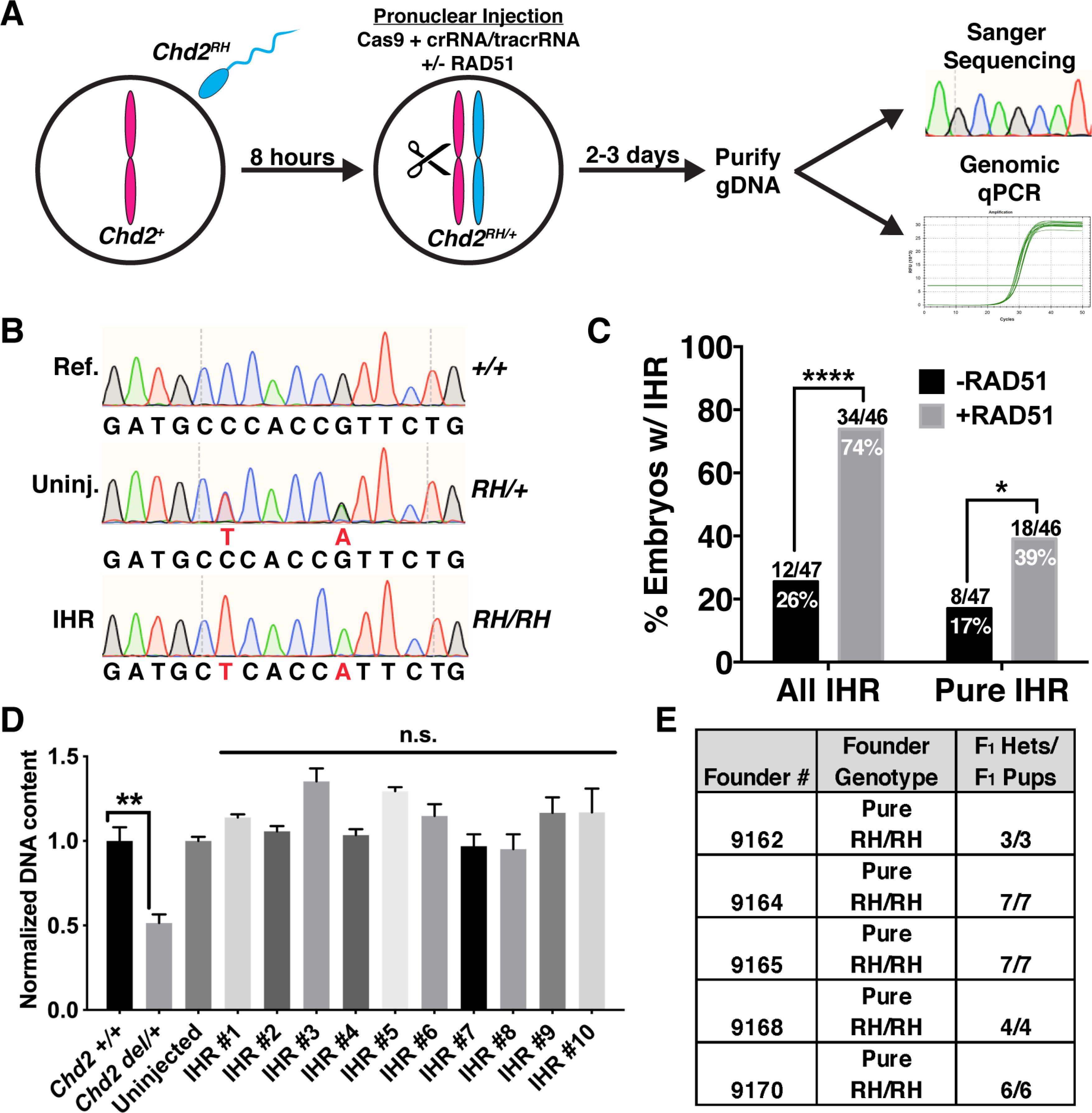
(A) Schematic of strategy for testing evaluating embryonic IHR. Wild-type oocytes were fertilized in vitro by sperm collected from Chd2RH/RH males and cultured for ~8 hours. PNI was then performed with Cas9 protein, tracrRNA, and crRNA specifically targeting the wild-type maternal allele. Injected embryos were then cultured for 2–3 days and collected at the morula or blastocyst stage. Half of the purified DNA was used for nested PCR and Sanger sequencing and the other half was used for multiplex PCR and subsequent qPCR to analyze genomic copy number at the Chd2 editing locus. (B) Representative chromatograms showing the wild-type reference sequence (Ref., top), an uninjected Chd2RH/+ embryo generated by IVF (Uninj., middle), and a pure Chd2RH/RH homozygous mutant (IHR, bottom) generated through IHR. (C) Quantification of Sanger sequencing results from IVF-derived embryos edited with or without RAD51. ‘All IHR’ includes mosaic embryos and was determined by identifying embryos with ≥3:2 ratio of R1684H allele to all other alleles (one-sided Fisher’s exact test). Pure IHR denotes embryos without mosaicism (p=0.018, one-tailed chi-square test). (D) Genomic qPCR targeting the Chd2RH locus in embryos carrying a large heterozygous deletion (Chd2del/+) as a control for copy-number sensitivity (lanes 1–2, unpaired t-test, n=3 embryos, error bars=SEM). Lanes 3–13 show genomic qPCR for the Chd2RH locus using DNA from an uninjected embryo and 10 randomly selected pure homozygous Chd2RH embryos (p>0.05, unpaired t-test, n=4 technical replicates per sample, error bars=SEM). (E) F1 genotyping results of litters derived from crosses between wild-type mice and 5 pure Chd2RH/RH F0 animals generated by the strategy described in (A), including RAD51. *p<0.05, **p<0.01, ****p<0.0001
