Figure 1: Identifying the Early Marker of Reprogramming.
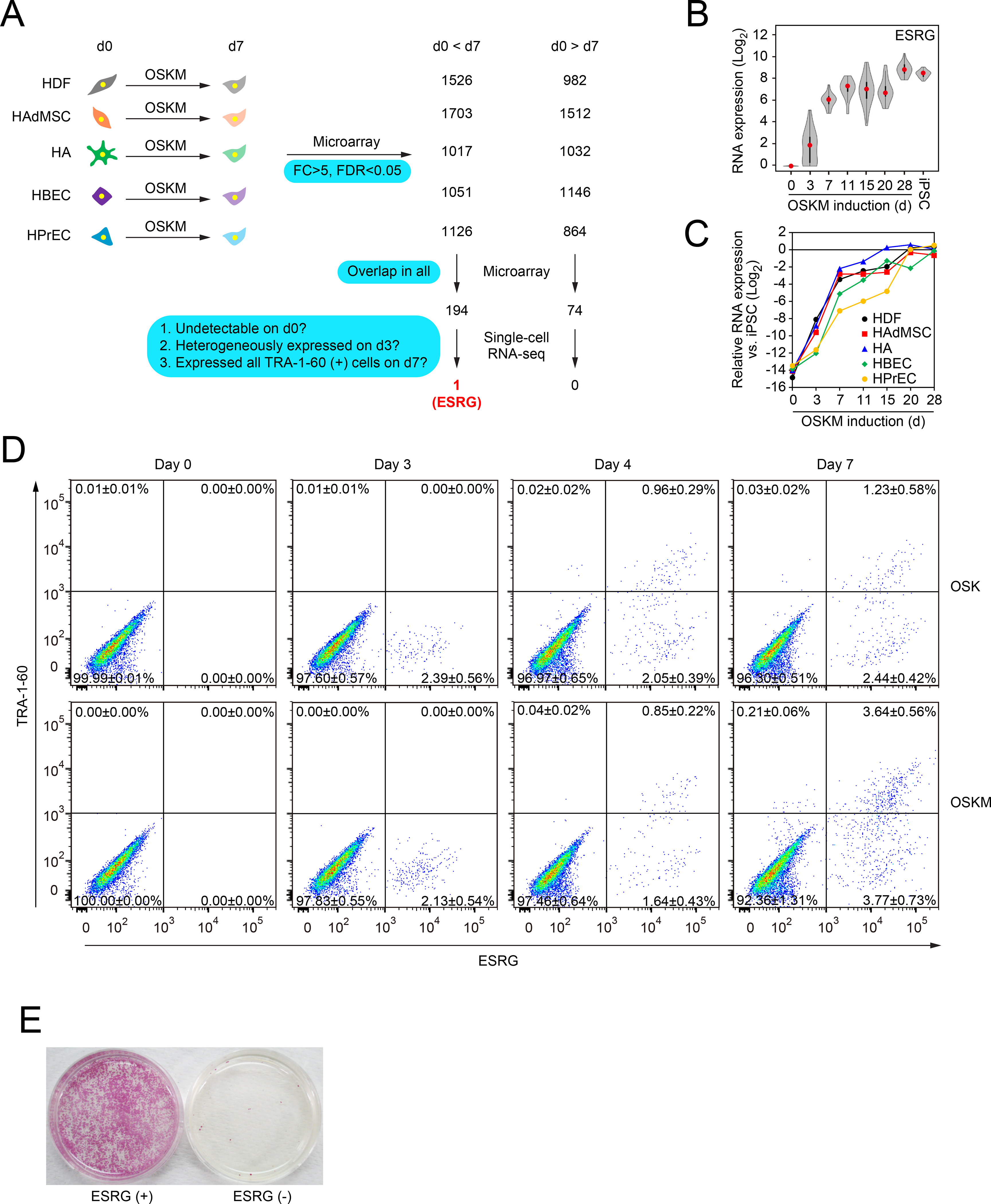
A, Extraction of commonly changed genes and narrowing down the candidate of reprogramming marker. We used microarrays to compare global gene expression of somatic cells (HDFs, HAdMSCs, HAs, HBECs and HPrECs) on day 0 (d0) and their TRA-1–60 (+) progenies on day 7 (d7) post-transduction of OSKM. n=3. By comparing differentially expressed genes between parental cells and TRA-1–60 (+) cells among five cell types, we obtained the indicated common genes. These commonly changed genes were evaluated by single-cell RNA-seq to narrow down the candidates. Blue boxes show the criteria of analyses. See also Tables S1 and S2.
B, Expression of ESRG during reprogramming. Shown are the results of expressing ESRG in parental HDFs (d0), OSKM-expressing cells on day 3 (d3), TRA-1–60 (+) cells at the indicated time points (d7–28), and iPSCs analyzed by single-cell RNA-seq. Twenty-four single cells were analyzed for each time point. Red dots indicate median values. Gray hour-glass shapes represent the distribution of RPKM + 1 value. See also Figure S1.
C, Expression of ESRG during reprogramming. Shown is the expression of ESRG in parental cells (d0), OSKM-expressing cells on day 3 (d3), TRA-1–60 (+) cells at the indicated time points (d7–28) derived from HDFs, HAdMSCs, HAs, HBECs and HPrECs analyzed by microarray. n=3.
D, TRA-1–60 (+) cells emerged only from ESRG (+) cells. Shown are representative histograms of ESRG-Clover (x-axis) and TRA-1–60 (y-axis) expression in ESRG-Clover fibroblasts (day 0) and those on days 3, 5 and 7 post-transduction by OSK or OSKM. n=3. See also Figure S1
E, ESRG (+) cells are origins of iPSCs. We introduced OSKM into ESRG-Clover fibroblasts by retroviral transduction and purified ESRG (+) and (−) cells by cell sorting. Sorted cells (100,000) were plated onto MMC-treated SNL feeder layers. A terminal stain was performed with red alkaline phosphatase on day 24. n=3.
