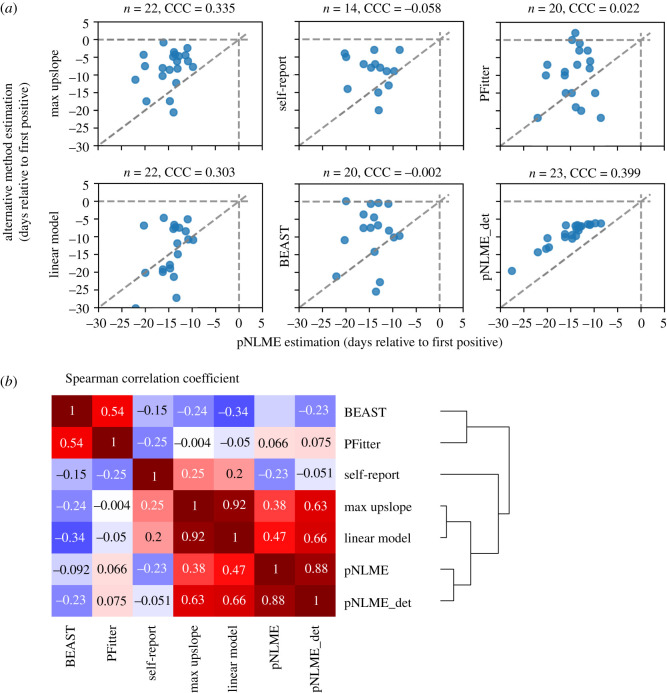
Figure 6.
Comparisons of the pNLME approach for infection timing versus five other methods. Methods include the maximum slope of any two points on the upslope, the best log-linear regression slope (linear model), self-report from trial participants, Bayesian phylogenetic inference of the median time to most recent common ancestor (BEAST), Poisson fitter (PFitter) diversity-based estimator and our approach restricted to only the deterministic (_det) component. (a) Best estimate of each available individual from each method expressed as predicted infection time relative to first positive viral load (all estimates are tabulated in electronic supplementary material, table S4). Comparison sample size n is denoted above each panel because some approaches are constrained by features of the data (e.g. detection of upslope, sequencing characteristics). Concordance correlation coefficients (CCC) show that individual agreement is generally weak: CCC = 1 when all data lie on the line y = x (dashed diagonal line). (b) Hierarchical clustering by the Spearman correlation illustrates which methods are predictive of others.