Figure 1.
Structure-based network analysis of the SARS-CoV-2 proteome identifies amino acid residues conserved in lineage B and C coronaviruses
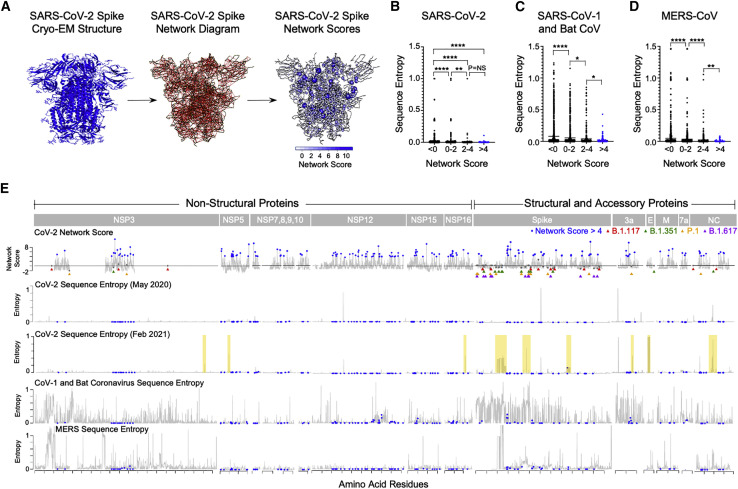
(A) Structure-based network analysis schematic for closed spike trimer (PDB: 6VXX) with amino acid residues (nodes) and non-covalent interactions (edges). Edge width indicates interaction strength and node size indicates relative network scores.
(B–D) Comparison of SARS-CoV-2 amino acid network scores (binned by network score: <0, 0–2, 2–4, and >4) with sequence entropy for SARS-CoV-2, sarbecoviruses (SARS-CoV-1/bat CoV), and MERS. (E) Alignment of SARS-CoV-2 network scores with sequence entropy values for SARS-CoV-2 in May 2020 and February 2021, sarbecoviruses (SARS-CoV-1/bat CoV) and MERS-CoV. Residues in blue indicate those with network scores >4. Network scores of residues mutated in the B.1.1.7 alpha (red triangles), B.1.351 beta (green triangles), P.1 gamma (yellow triangles), and B.1.617.2 delta variant (purple triangles) are depicted in gray. Yellow boxes indicate new areas of sequence variation in SARS-CoV-2 that emerged between May 2020 and February 2021. Statistical comparisons were made using Mann-Whitney U test. For comparisons of more than two groups, Kruskal-Wallis test with Dunn’s pos hoc analyses was used. Calculated p values were as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.