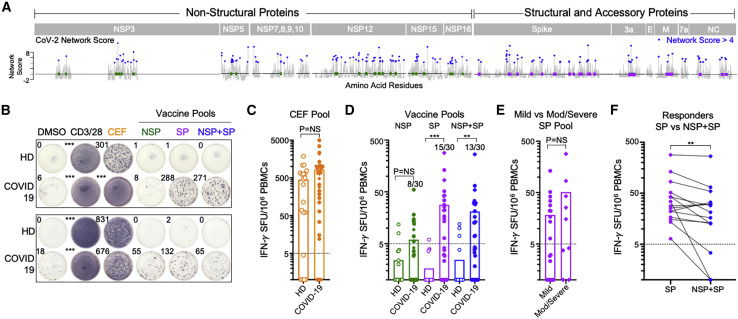
Figure 4.
CD8+ T cells from convalescent COVID-19 individuals recognize highly networked epitopes derived from structural and accessory proteins
(A) Location of highly networked HLA-stabilizing CD8+ T cell epitopes in non-structural proteins (NSPs; green) and structural proteins (SPs; purple) across the SARS-CoV-2 proteome.
(B) Representative IFN-γ ELISpot data for two pairs of healthy donors (HDs) and COVID-19 patients following incubation with DMSO, anti-CD3/CD28 antibodies, the CEF peptide pool, the highly networked NSP peptide pool (n = 56), the highly networked SP peptide pool (n = 53), and the combined NSP + SP peptide pool (n = 109).
(C) Magnitude of IFN-γ+ CD8+ T cell responses to the CEF peptide pool in HDs (open, n = 20) and COVID-19 patients (filled, n = 30). Mild (filled circles, n = 21) and moderate-to-severe COVID-19 patients (filled diamonds, n = 9) are denoted.
(D) Magnitude of IFN-γ+ CD8+ T cell responses to the highly networked SARS-CoV-2 NSP epitope pool (green), the SP epitope pool (purple), and the combined NSP + SP epitope pool (blue) in HDs (open, n = 20) and COVID-19 patients (filled, n = 30). The number of positive responders relative to the total number of individuals analyzed is indicated.
(E) Magnitude of IFN-γ+ CD8+ T cell responses against the highly networked SARS-CoV-2 SP epitope pool (purple) in mild (n = 21) and moderate-to-severe COVID-19 patients (n = 9).
(F) Comparison of the magnitude of IFN-γ+ CD8+ T cell responses to SP and NSP + SP peptide pools in COVID-19 SP peptide pool responders (n = 15).
Statistical comparison was made using a Wilcoxon matched-pairs test. All other statistical comparisons were made using a Mann-Whitney U test. Calculated p values were as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Table 1.