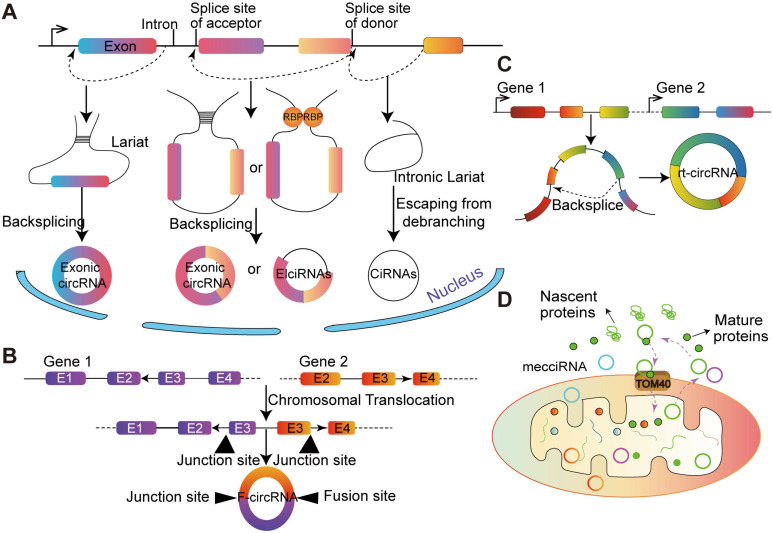
Figure 1.
The biogenesis of circRNAs and special types of circRNA. (A) Lariat-driven circularization. Exon skipping events result in lariat structure, then the lariat is removed, and finally circRNA is formed. Intron-pairing-driven circularization. There are complementary sequence motifs between the introns sequences flanking the exon the base complementary forms a circular structure and the introns are removed or retained to form ecircRNAs or EIciRNAs. RBPs-pairing-driven circularization. The RBP interaction on the exon flanking sequence promotes the formation of circRNAs. CiRNAs. The ciRNAs were obtained from intronic lariat escape from debranching. (B) F-circRNA. CircRNA derived from the back-splicing of fusion-gene. (C) Rt-circRNA. Stop-codon read-through from read-through circRNA rt-circRNA. (D) MecciRNAs. CircRNAs encoded by the mitochondrial genome.