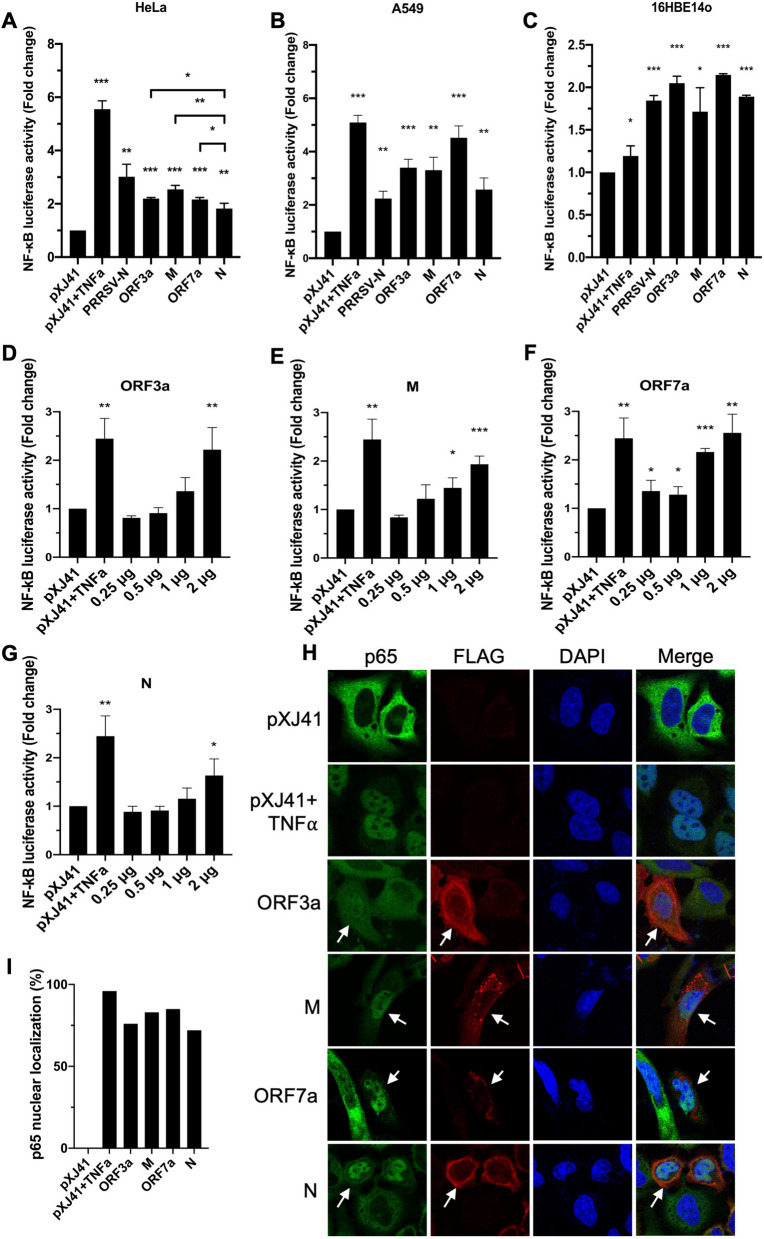
Figure 3.
Cellular NF-κB response mediated by SARS-CoV-2 proteins in three different cell types. HeLa cells (A,D–G), A549 cells (B), or 16HBE14o cells (C) were co-transfected with pNF-κB-Luc (0.5 μg), pRL-TK (0.05 μg), and each of indicated SARS-CoV-2 genes. At 24 h post-transfection, cells were incubated with TNF-α (20 ng/ml) for 6 h, and cell lysates were prepared for luciferase assays. Luciferase activities were measured using the Dual Luciferase assay system according to the manufacturer’s instructions (Promega). Relative luciferase activities were obtained by normalizing the firefly luciferase to Renilla luciferase activities. Values of the relative luciferase activity in the pXJ41 control group were set as 1, and the values for individual viral proteins were normalized using that of the pXJ41 control. Error bars mean ± standard deviation (s.d.). (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. (H) HeLa cells were transfected with 2 μg of indicated genes for 24 h and treated or mock-treated with TNF-α (20 ng/ml) for 30 min. The cells were stained with α-p65 Mab (green) and α-FLAG PAb (red). Nuclei (blue) were stained with DAPI. White arrows indicate viral protein-expressing cells. (I) The percentages of cells showing p65 nuclear localization were calculated using the following formula: (Number of cells showing p65 nuclear staining)/(50 cells expressing viral protein) × 100.