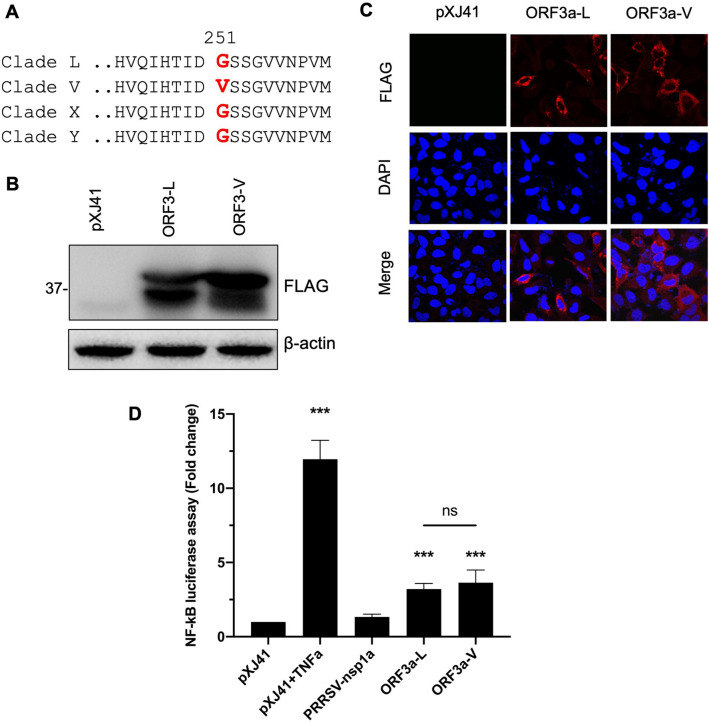
Figure 6.
NF-κB activation by ORF3a from different clades of SARS-CoV-2. (A) Sequence alignments of four major clades of SARS-CoV-2 ORF3a. Single amino acid change (G251V) was identified in clade V. ORF3a genes from clade L and V were fused with the FLAG-tag and cloned in the pXJ41 expression vector and designated as ORF3a-L and ORF3a-V. Using α-FLAG PAb, expression patterns of ORF3a-L and ORF3a-V were demonstrated by immunoblot (B) or IFA (C). Beta-actin served as a loading control. The full-length blot of (B) is presented in Supplementary Fig. S2. (D) Cells were co-transfected with pNF-κB-Luciferase (0.5 μg), pRL-TK (0.05 μg), and each (0.5 μg) of indicated SARS-CoV-2 ORF3a genes for 24 h. Cells were treated or mock-treated with TNF-α (20 ng/ml) for 6 h, and cell lysates were used for luciferase assays. Relative luciferase activities were obtained by normalizing the firefly luciferase to Renilla luciferase activities. Values of the relative luciferase activity in the pXJ41 control group were set as 1, and the values for individual viral proteins were normalized using that of the pXJ41 control. Error bars mean ± standard deviation (s.d.). (n = 3). ns non-significance (P > 0.05), ***P < 0.001.