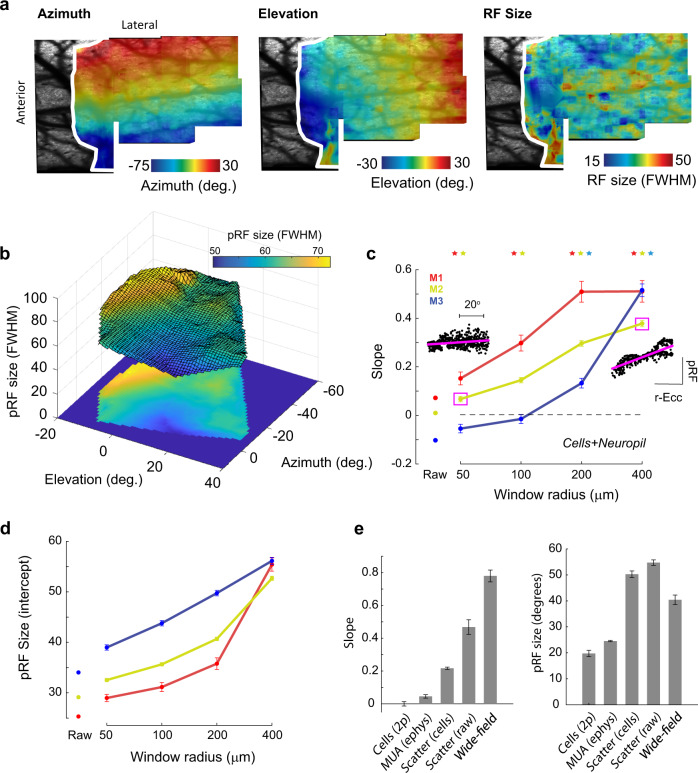
Fig. 5. pRFs generated from raw two-photon images show stronger scattering.
a Example maps of azimuth and elevation generated from the (smoothed) raw two-photon images show a clear retinotopic organization in agreement with the RFs measured for the individual cells. The white border indicates the boundary of V1 as determined by field-sign analysis (Methods). The map of RF size (right panel) showed no clear organization at this level of spatial detail. These images form the input into the scatter analysis. b Measures of pRF size obtained from performing the scatter analysis on the retinotopic maps shown in (a), a window size of 400 μm was used to generate this image. The smallest pRFs are in the region representing the focea. c The slope of the regression of pRF size on r-eccentricity for the raw image data approached the values from the wide-field data in Fig. 1i for the larger analysis windows. Example regression fits for mouse M2 are shown in the insets. Asterisks mark significant values, p < 0.05, t-test, two-tailed, and the error bars indicate 1 SEM. d The intercept term of the regression gives the expected pRF size at the focea. This approached the minimum values observed in the wide-field data (approximately 40–50°) only at window sizes of 200–400 μm radius, suggesting that windows of this size best capture the signals that are measured in the wide-field data. The pattern was consistent across mice (colored lines). Error bars indicate 1 SEM. e Summary of the slope (left) and intercept (values) for the different techniques. The results indicate that the small pRFs in the focea are caused by reduced the scatter of RFs across cells. Techniques that measured individual cells or small multi-units (electrophysiology, two-photon cell analysis) did not find strong relationships, whereas those that measured activity pooled over many cells (two-photon scatter analyses and wide-field analysis) found a relationship. The data from the cell+neuropil analysis (i.e., raw) had slope values closest to the wide-field data. The values for the scatter analyses are taken from the 400 μm radius analysis windows. Error bars indicate 1 SEM across animals.