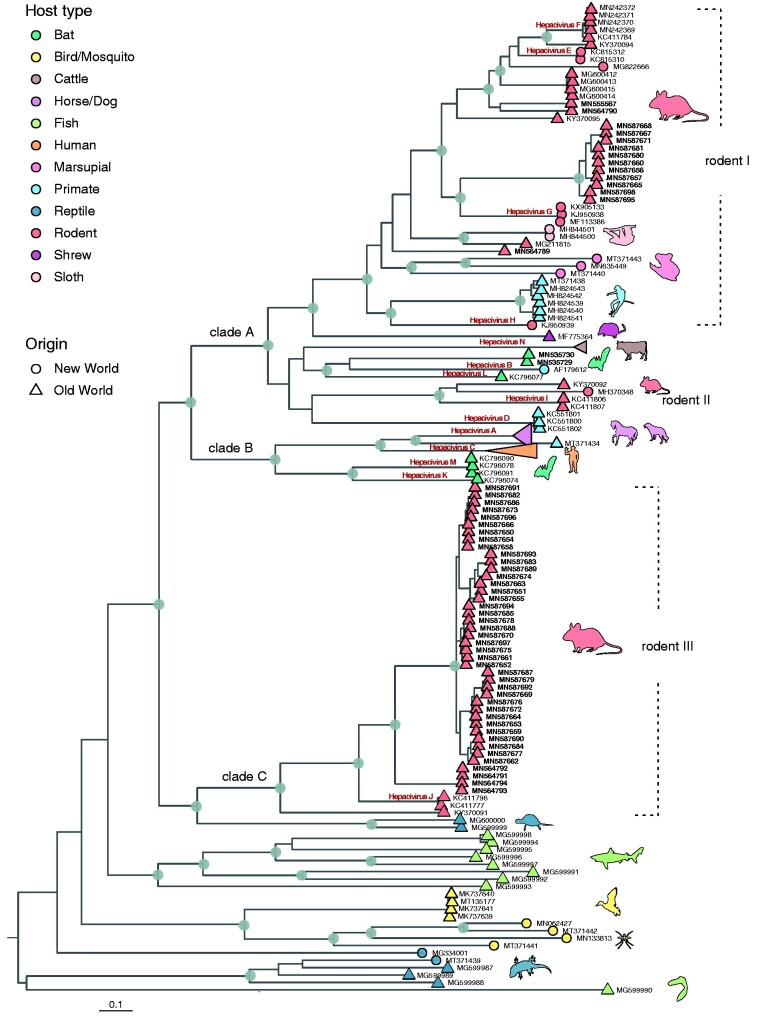
Figure 2.
Genome-wide phylogenetic reconstruction of hepaciviruses. ML tree of all available (n = 130) and novel (n = 58) hepacivirus genomes. Silhouettes indicate hosts and are coloured according to their broader host type: bats (green), birds and mosquito (yellow), cattles (brown), equids and dog (lilac), fishes (lime green), humans (peach orange), marsupials (pastel pink), primates (light blue), reptiles (steel blue), rodents (salmon), shrew (plum) and sloths (champagne pink). Grey circles indicate internal nodes with bootstrap support , while designated virus species names are shown in bold bordeaux text. Circles at the tips denote New World origin, while triangles represent the Old Wolrd viruses. Novel genomes generated in this study are labeled with bold text. Clades A, B and C have been provisionally named for the purpose of discussing the mammalian hepacivirus lineages in the main text.