Abstract
The appearance of new variants of SARS‐CoV‐2 has recently challenged public health authorities with respect to tracking transmission and mitigating the impact in the evolving pandemic across countries. B.1.525 is considered a variant under investigation since it carries specific genetic signatures present in P.1, B.1.1.7, and B.1.351. Here we report genomic evidence of the first likely imported case of the SARS‐CoV‐2 B.1.525 variant, isolated in a traveler returning from Nigeria.
Keywords: B.1.525 variant, Brazil, genomic surveillance, SARS‐CoV‐2
1. INTRODUCTION
Described for the first time in mid‐December 2020, the SARS‐CoV‐2 variant B.1.525 is currently classified as a variant under investigation (VUI) due to the presence of a notable group of missense mutations believed to be of particular importance due to their potential for increased transmissibility, virulence, and reduced effectiveness of vaccines. 1 As of April 15, 2021, B.1.525 had already been reported in 48 countries across Europe, North America, Africa, Oceania, and Asia and the most affected countries are the United States of America, United Kingdom, Germany and Nigeria, with 19.36%, 19.74%, 10.49%, and 6.70% of detected cases, respectively. 2 In the latter country, at the beginning of February 2020, the B.1.525 variant was identified in 45% of all sequenced samples. 2 Also known as 20A/S:484K (https://covariants.org/variants/20A.S.484K) and VUI 202102/03 (https://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-cases-data), the B.1.525 variant harbors some genetic signatures related to diverse putative effects on viral fitness, which are shared by other variants of concern (VOC), highlighting the potential impact of dissemination in countries where it has not yet been detected: (i) Q677H—described as modulating transmissibility; (ii) (Δ144)—associated with immune escape; (iii) Δ106‐108—detected already in B.1.1.7, P.1, and B.1.351; (iv) E484K—present in 501.V2, P1, and P2 variants; (v) N439K—found in Y453F, B.1.141, B.1.258 variants; and (vi) two deletions ΔH69/ΔV70 detected in B.1.1.7. 3
In an effort to quickly detect and characterize all circulating SARS‐CoV‐2 variants in the state of Bahia, located in northeastern Brazil, public health authorities have enhanced epidemiological and genomic investigation by screening individuals who report travelling from countries where variants of interest and/or concern are circulating. Here, we report genomic evidence of the first likely imported case of the SARS‐CoV‐2 B.1.525 variant, isolated in a traveler returning from Nigeria.
A 74‐year‐old female nun, resident in Feira de Santana, Bahia state (Northeast Brazil), reported symptoms compatible with viral infection on February 10th, 2021. She reported travel history from Africa (Nigeria and Ethiopia countries). The patient reported fever, cough, asthenia, prostration, chills, loss of appetite and headache for approximately 10 days. The patient procured care at the São Matheus hospital in Feira de Santana and she was submitted to paracetamol 750 mg for 7 days. However, since disease was classified as mild, she was treated at home, not requiring hospitalization.
2. MATERIALS AND METHODS
Viral RNA was extracted from the nasopharyngeal swab sample and submitted to multiplex real‐time PCR using the Allplex™ 2019‐nCoV Assay (Seegene). The RT‐PCR–positive sample was then submitted to viral genomic amplification and posterior sequencing using theIon GeneStudio™ S5 Plus System (Life Technologies) in conformity with the manufacturer's instructions. A consensus sequence was generated by de novo assembling using the Genome Detective tool (https://www.genomedetective.com/). 4 Lineage assignment was performed using the Pangolin lineage classification software tool. 5
The newly identified B.1.525 isolate was compared to a diverse pool of genome sequences (n = 3852) sampled worldwide collected up to March 21, 2021. All sequences were aligned using the MAFFT tool, 6 and IQ‐TREE 7 was used for phylogenetic analysis using the maximum likelihood approach. 8 The mutation pattern of the VUI was analyzed using the NextClade online tool. 9
3. RESULTS
The cycle threshold (Ct) values of the four targets of the SARS‐CoV‐2 assay were: RNA‐dependent RNA polymerase (RdRp)—30.97; envelope protein (E)—29.65; nucleocapsid protein (N)—29.05; and internal control (IC)—28.70. Viral genomic RNA amplification and sequencing produced a total of 1,417,554 mapped reads, with corresponding coverage of 99.6% and a mean sequencing depth of 8901.4X. The newly generated whole genome sequence was assigned as the B.1.525 variant in conformity with Pangolin lineage classification.
To accurately establish evolutionary relationships among the newly generated sequence and other isolates of SARS‐CoV‐2, we then subjected a combined data set to phylogenetic inference. Analysis of the phylogenetic tree indicated that the VUI (BA125) clustered within a clade containing other sequences previously assigned as the B.1.525 variant (Figure 1A,B) with bootstrap value of 100. Using NextClade, the mutation pattern of BA125 (Figure 1C) was determined, and no newly acquired mutations were identified.
Figure 1.
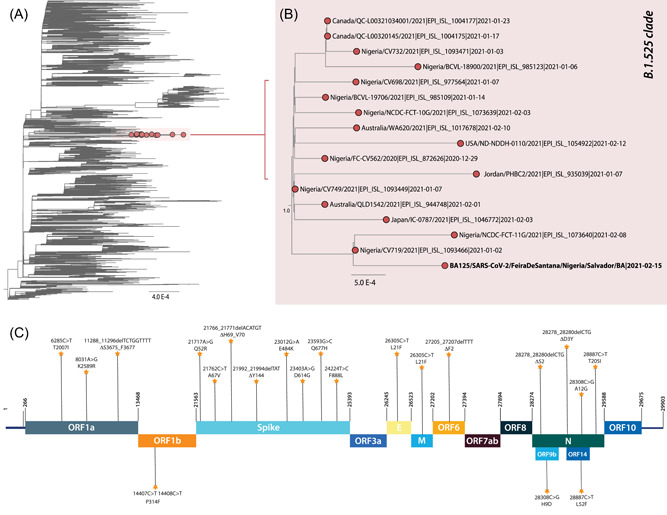
Genomic detection of the SARS‐CoV‐2 B.1.525 variant of interest in Bahia state, Northeast Brazil. (A) ML phylogenetic tree including the newly B.1.525 isolateobtained in this study plus n = 3852 representative SARS‐CoV‐2 strains collected up to March 21st, 2021. (C) Representation of the zoom of the B.1.525 clade. Branch support (Bootstrap and SH‐aLTR >0.9) is shown at key nodes. (D) Variant maps of the B.1.525 lineage‐defining‐mutations mapped against the SARS‐CoV‐2 genome structure. Most common mutations are highlighted. ML, maximum likelihood
4. DISCUSSION
Here we report the early detection of the B.1.525 variant in northeastern Brazil (state of Bahia), highlighting the continuing need for genomic and epidemiological surveillance to detect variants previously not yet identified in this state. This emergent lineage, carries S: Q677H, the F888L in addition to several mutations seen in B.1.1.7 (501Y.V1) and other VOCs (P.1 and B.1.351), such as S: del 69‐70 and S: del 144, and also, S: E484K related to increased risk of disease severity, risk of vaccine escape10, 11 and higher transmissibility. This emerging VUI appears to differ from all other variants by having both the E484K‐mutation and a new F888L mutation (a substitution of phenylalanine (F) with leucine (L) in the S2 domain of the spike protein), that may have functional importance. Considering also the recent concern of the rapid rise 12 of this novel variant worldwide and considering the constellation of mutation into the S protein, our findings reinforce the need for active monitoring of travelers, to follow the real‐time spread of new SARS‐CoV‐2 variants with possible implications for public health policies and immunization strategies.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ETHICAL STATEMENT
This study was approved by the Ethics Review Committee of the Federal University of Minas Gerais (CEP/CAAE: 32912820.6.1001.5149 approval number).
AUTHOR CONTRIBUTION STATEMENT
Conception and design: FP, ST, and MG; Data collection: MML, LROS, VBN, MKAG, JGL; Investigations: FP, ST, MG, VF and LCJA; Data Analysis: FP, ST, MG, VF and LCJA; Writing –Original: FP, ST, MG; Draft Preparation: FP, ST, VF, LCJA and MG; Revision: TO, JL, AL, LCJA and MG.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
This study was financed by Laboratório Central de Saúde Pública da Bahia (LACEN‐BA) and Secretaria da Saúde do Estado da Bahia (SESAB) and supported by Pan American Health Organization PAHO/WHO. We also thank Epidemiological Surveillance Division of the Municipal Health Department of Feira de Santana, Bahia. ST is supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001. MG is supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). We also would like to thank all the authors who have kindly deposited and shared genome data on GISAID. A table with genome sequence acknowledgments can be found in Table S1.
Pereira F, Tosta S, Lima MM, et al. Genomic surveillance activities unveil the introduction of the SARS‐CoV‐2 B.1.525 variant of interest in Brazil: Case report. J Med Virol. 2021;93:5523–5526. 10.1002/jmv.27086
Felicidade Pereira and Stephane Tosta contributed equally to this work.
Contributor Information
Luiz C. Alcantara, Jr., Email: luiz.alcantara@ioc.fiocruz.br.
Marta Giovanetti, Email: giovanetti.marta@gmail.com.
DATA AVAILABILITY STATEMENT
Newly generated SARS‐CoV‐2 sequence have been deposited in GISAID under accession number EPI_ISL_1583653.
REFERENCES
- 1. Centers for Disease Control and Prevention . “Emerging SARS‐CoV‐2 Variants.” www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html. [PubMed]
- 2. Elbe S, Buckland‐Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Challenges. 2017;1:33‐46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Public Health England . “SARS‐CoV‐2 variants of concern and variants under investigation in England”. Technical briefing 7. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/972247/Variants_of_Concern_VOC_Technical_Briefing_7_England.pdf
- 4. Vilsker M, Moosa Y, Nooij S, et al. Genome detective: An automated system for virus identification from high‐throughput sequencing data. Bioinformatics. 2019;35(5):871‐873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Toole Á, Scher E, Underwood A, et al. Pangolin: lineage assignment in an emerging pandemic as an epidemiological tool. https://github.com/cov-lineages/pangolin [DOI] [PMC free article] [PubMed]
- 6. Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large‐scale multiple sequence alignments. Bioinformatics. 2018;34:2490‐2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nguyen L‐T, Schmidt HA, von Haeseler A, Minh BQ. IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol. 2015;32:268‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sagulenko P, Puller V, Neher RA. TreeTime: maximum‐likelihood phylodynamic analysis. Virus Evol. 2018;4:1‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nextclade . Nextstrain tool Nextclade beta version 0.4.9. https://clades.nextstrain.org/
- 10. Wang WB, Liang Y, Jin YQ, Zhang J, Su JG, Li QM. E484K mutation in SARS‐CoV‐2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: Binding free energy calculation studies. bioRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO . COVID‐19 weekly epidemiological update. Data as received by WHO from national authorities, as of 11 April 2021, 10 am CET.WHO. https://apps.who.int/iris/bitstream/handle/10665/340826/nCoV-weekly-sitrep13Apr21-eng.pdf?sequence=1
- 12. Lineage B.1.525 . https://cov-lineages.org/lineages/lineage_B.1.525.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
Newly generated SARS‐CoV‐2 sequence have been deposited in GISAID under accession number EPI_ISL_1583653.


