Abstract
Objectives
To describe the first outbreak of Candida auris in Brazil, including epidemiological, clinical and microbiological data.
Methods
After the first Candida auris‐colonised patient was diagnosed in a COVID‐19 ICU at a hospital in Salvador, Brazil, a multidisciplinary team conducted a local C. auris prevalence investigation. Screening cultures for C. auris were collected from patients, healthcare workers and inanimate surfaces. Risk factors for C. auris colonisation were evaluated, and the fungemia episodes that occurred after the investigation were also analysed and described. Antifungal susceptibility of the C. auris isolates was determined, and they were genotyped with microsatellite analysis.
Results
Among body swabs collected from 47 patients, eight (n = 8/47, 17%) samples from the axillae were positive for C. auris. Among samples collected from inanimate surfaces, digital thermometers had the highest rate of positive cultures (n = 8/47, 17%). Antifungal susceptibility testing showed MICs of 0.5 to 1 mg/L for AMB, 0.03 to 0.06 mg/L for voriconazole, 2 to 4 mg/L for fluconazole and 0.03 to 0.06 mg/L for anidulafungin. Microsatellite analysis revealed that all C. auris isolates belong to the South Asian clade (Clade I) and had different genotypes. In multivariate analysis, having a colonised digital thermometer was the only independent risk factor associated with C. auris colonisation. Three episodes of C. auris fungemia occurred after the investigation, with 30‐day attributable mortality of 33.3%.
Conclusions
Emergence of C. auris in Salvador, Brazil, may be related to local C. auris clade I closely related genotypes. Contaminated axillary monitoring thermometers may facilitate the dissemination of C. auris reinforcing the concept that these reusable devices should be carefully cleaned with an effective disinfectant or replaced by other temperature monitoring methods.
Keywords: Candida auris, COVID‐19, intensive care, Brazil
1. INTRODUCTION
In the last decade, we have witnessed the emergence and worldwide nosocomial spread of the new human opportunistic pathogen Candida auris. 1 , 2 Outbreaks of hospital‐acquired infections and the potential of C. auris to develop multidrug resistance have alarmed the medical and scientific communities. 3 Previous C. auris‐free hospitals now have this yeast among the leading agents of bloodstream infections. 4 , 5 , 6
During the ongoing COVID‐19 pandemic, the overwhelmed intensive care units (ICUs) have been a fertile ground for the emergence and spread of C. auris. 7 , 8 , 9 Until end of 2020, C. auris was not reported in Brazil 10 but recently we reported the first two patients that had been hospitalised due to severe COVID‐19. 11 After being notified by the local hospital infection control team (HICT), the Brazilian Ministry of Health and the National Sanitary Surveillance Agency (NSSA) set in place a task force to map and control a possible outbreak.
Based on published guidelines 12 , 13 , 14 and on previous reported experiences, 15 , 16 , 17 an intervention took place, including cohorting and collection of surveillance cultures of potential C. auris‐colonised patients. Details about the cross‐sectional investigation of the first outbreak of C. auris in Brazil and the fungemia episodes that occurred after the investigation are described.
2. MATERIAL AND METHODS
2.1. Settings and definitions
The outbreak took place in a 330‐bed hospital in Salvador, Bahia, Brazil. The hospital has three intensive care units (ICUs) with a total of 66 beds for critically ill patients. The hospital has an additional 63 semi‐intensive care unit (SICU) and 201 ward beds. In March 2020, when the SARS‐CoV‐2 pandemic began in Brazil, one of the three ICUs with 20 beds were allocated to patients with severe COVID‐19. On December 2, 2020, the first Candida auris‐colonised patient was diagnosed at the hospital. 11 The patient had spent 38 days at the COVID‐19 ICU, and after a negative nasopharyngeal PCR for SARS‐CoV‐2, was transferred to one of the three SICU.
The cross‐sectional investigation took place on December 16, 2020. All patients were or had been hospitalised at the COVID‐19 ICU, as well as their close contacts (defined as patients that were hospitalised at the same unit and attended by the same healthcare worker team) were considered potential C. auris‐colonised patients and were investigated. Fungemia episodes that occurred after the investigation were also analysed.
Since no invasive procedures were required for the investigation and due to the urgent sanitary relevance, the Brazilian Ministry of Health and the institutional review board waved patients’ and healthcare workers’ consent to carry on the study. Patient and healthcare worker anonymity was assured during this investigation.
2.2. Screening cultures for Candida auris of patients and healthcare workers
Candida auris screening cultures with sterile swabs (one per site, premoistened with sterile saline) were used to collect samples from the following sites of the suspected colonised patients: axillae, groins, ears, nostrils and pressure ulcers when present. Additionally, healthcare workers underwent a visual inspection of their hands, and skin or trophic nail lesions were also swabbed. After sampling, the swabs were immediately inoculated in 15‐ml conical tubes (one swab per tube), containing Sabouraud dextrose broth (SDA) enriched with 10% NaCl, and shipped to the laboratory to be incubated at 40°C for 7 days. Tubes were checked every 24 h for yeast growth, and positive samples were plated on chromogenic agar (CHROMagar™ Candida, DIFCO) that were incubated for 24–48 h at 37°C.
2.3. Environmental screening
Samples were collected with 3M sponge‐sticks (Fisher Scientific, Pittsburg, Pennsylvania) as recommended. 12 , 18 The environment from the index case and from the potential C. auris‐colonised patients at the ICUs or SICUs had an initial visual inspection, and the high‐touch inanimate sites were selected to be sampled (one sponge stick per site): bed rails; mechanical ventilators, vital signs monitors and intravenous infusion pumps (composite sample); reusable digital thermometers (inside plastic recipient on the wall); and tray tables. Additional areas in the vicinity of the patients, including faucets, sinks, computer keyboards (protected with plastic film) and mouse, ultrasound probes and hand sanitiser wall dispensers were also sampled. Immediately after sampling, the sponge‐sticks were put into sterile plastic bags, sealed and shipped to the laboratory. In the laboratory, the sponge‐sticks were inserted into 50‐ml conical tubes (one sponge‐sticks per tube) containing SDB with 10% NaCl and incubated at 40°C for 7 days. Tubes were checked every 24 h for yeast growth, and positive samples were plated on chromogenic agar (CHROMagar™ Candida) and incubated at 37°C for 24–48 h.
2.4. Blood, urine and central venous catheter tip cultures
Clinical cultures were collected from patients during sepsis investigation. Blood cultures were carried out with BacT/ALERT aerobic bottles (bioMérieux, Marcy‐l'Etoile, France) and incubated in the automated BacT/ALERT 3D system (bioMérieux) at 35°C. Central venous catheter tip (CVC‐tip) samples and positive blood culture samples were inoculated on blood sheep agar that were incubated for 24–48 h at 37°C. Urine cultures were plated on chromogenic medium for urinary samples and incubated 24–48 h at 37°C.
2.5. Species identification
Yeast colonies from blood cultures, CVC‐tip or urinary samples were initially identified by the Vitek 2 system (YST cards, bioMérieux). Clinical isolates with C. auris identified by the Vitek 2 system (bioMérieux), and yeasts recovered from screening cultures were identified by MALDI‐TOF mass spectrometry (VitekMS, bioMérieux, Marcy‐l'Etoile, France). Final species characterisation was carried out by ITS rDNA sequencing analysis. 19 All sequences were deposited and are available at GenBank (Supplementary Material, https://www.ncbi.nlm.nih.gov/genbank/).
2.6. Antifungal susceptibility testing
The in vitro activity of amphotericin B (AMB, Sigma‐Aldrich, Saint Louis, MO, USA), fluconazole (Sigma‐Aldrich), voriconazole (Sigma‐Aldrich) and anidulafungin (Sigma‐Aldrich) against the C. auris isolates was evaluated by the CLSI broth microdilution reference method. 20 Plates were incubated at 37°C and minimal inhibitory concentrations (MICs) were read after 24 h.
2.7. Microsatellite typing
To investigate the clonality between the clinical and environmental C. auris isolates, microsatellite typing with four multiplex PCR reactions was (M2, M3‐I, M3‐II, M9) performed as previously described. 21 A selection of clinical and environmental Brazilian isolates, and representative strains from other countries were analysed. UPGMA dendrogram of short tandem repeats (STR) genotypes was constructed with the software BioNumerics, version 7.6.1 (Applied Maths NV‐bioMérieux, Sint‐Martens‐Latem, Belgium).
2.8. Epidemiological Investigation and statistical analyses
The investigators filled a form for all cases with clinical samples positive for C. auris and for all potentially colonised patients. The form required information regarding demographics; comorbidities and baseline diseases, including COVID‐19 diagnosis; associated conditions including invasive medical procedures; and previous exposure to antimicrobials, corticosteroids or antifungals. These data were further combined with the screening cultures results, and the patients were than finally classified as C. auris colonised (case) or non‐colonised (controls). Cases with missing clinical or microbiologic data were excluded.
To describe the potential risk factors for C. auris colonisation, data comparisons between the colonised vs non‐colonised patients were carried out with SPSS software v.22 (IBM, Armonk, NY, USA). Categorical variables were expressed as percentage and continuous variables as median ±standard deviation (SD). Differences between the groups were evaluated with Chi‐squared test, Fisher's exact test or the Mann‐Whitney U test. Variables associated with p values < .3 on univariate basis were introduced into the multivariate model. Two‐tailed p values < .05 were considered statistically significant.
To describe the bloodstream infection episodes, data regarding CVC removal, antifungal treatment, and 30‐day outcomes were also collected. The doctor in charge of the patient was asked to classify the patient's death as attributable or non‐attributable to the C. auris fungemia episode.
3. RESULTS
A total of 66 potentially C. auris‐colonised patients were identified, not including the first case of C. auris in that hospital (CVC‐tip culture), and another patient who died after having a C. auris‐positive urine culture was not eligible for culture screening evaluation. A total of 200 superficial body swabs were collected from 47 ICU/SICUs patients (first colonised patient and 46 potential colonised patients), generating eight (n = 8/47, 17%), five (5/47, 10.6%), three (3/47, 6.4%) and two (2/47, 4.3%) C. auris‐positive samples from the axillae, groins, nostrils and ears, respectively. All nine samples collected from pressure ulcers were C. auris‐negative. Three‐hundred and one healthcare workers had visual inspection of their hands, and five had skin or trophic nail lesions. None of these five healthcare workers had positive cultures for C. auris.
The environmental investigation led to the collection of 204 samples from the ICU/SICUs. Among the samples collected from inanimate surfaces, the digital thermometers had the highest rate of positive cultures (8/47, 17%), followed by bed rails (7/47, 14.9%), vital signs monitors/intravenous infusion pumps (5/47, 10.6%) and tray tables (5/47, 10.6%). Twelve samples collected from either the faucets, sinks, computer keyboards and mouse, ultrasound probes or hand sanitiser wall dispensers were negative for C. auris. Of note, one patient with an axillae positive sample had negative cultures from the surrounding inanimate surfaces, and two individual ICU rooms had positive cultures from the environment but the surveillance cultures from the patients were negative. Figure 1 summarises the results of the patients’ and environmental cultures from the ICUs and SICUs. The remaining 20 potentially colonised patients from the wards had negative axillae, groins, nostrils and ear swab cultures. From the 66 potentially colonised patients, eight (12.1%) were confirmed to carry C. auris. The clinical, epidemiological and microbiological details about the C. auris‐colonised patients are provided in Table 1.
FIGURE 1.
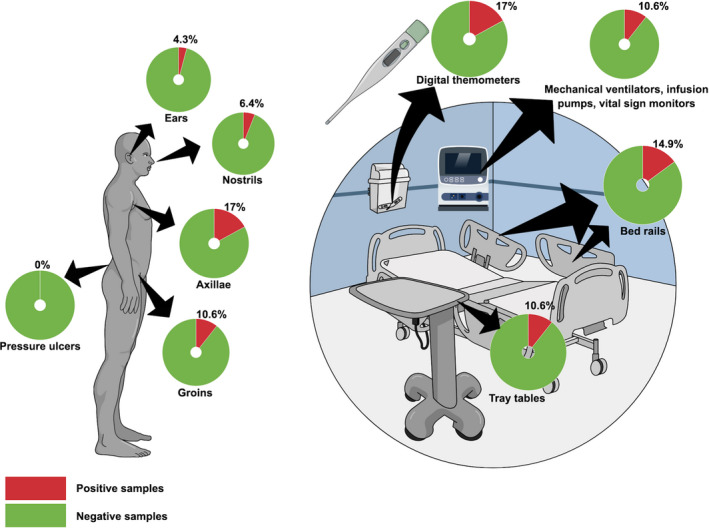
Illustration of results of Candida auris screening cultures among patients and different inanimate surfaces
TABLE 1.
Epidemiological and clinical data of the ten Candida auris‐colonised patients found during the investigation
| Age/sex | Motive of Admission | Baseline diseases | Admission to positive culture | Antimicrobial exposure | Antifungal exposure | Corticosteroid exposure | MV | CVC | Urinary catheter | HD | Culture sample |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 59/M a | Severe COVID‐19 | Leg deep venous thrombosis | 42 days | Yes | Yes | Yes | Yes | Yes | Yes | Yes | CVC‐tip |
| 79/M | Severe COVID‐19 | Biliary lithiasis | 46 days | Yes | Yes | Yes | Yes | Yes | Yes | No | CVC‐tip, axillae, groin, nostrils and ear swabs |
| 72/M b | Severe COVID‐19 | Stroke, dementia | 36 days | Yes | Yes | Yes | Yes | Yes | Yes | No | Urine |
| 68/M | Upper gastrointestinal bleeding | Hypertension, diabetes, chronic renal failure, obesity | 8 days | Yes | Yes | No | Yes | Yes | Yes | Yes | Axillae swab |
| 58/M | Femur fracture, severe COVID‐19 | Hypertension, diabetes, obesity | 27 days | Yes | No | Yes | No | Yes | No | No | Axillae, groins swabs |
| 63/M | Severe COVID‐19 | Hypertension, diabetes | 18 days | Yes | No | Yes | No | Yes | No | No | Axillae, groin and nostril swabs |
| 75/F | Severe COVID‐19 | Hypertension, diabetes, hypothyroidism | 32 days | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Axillae swab |
| 63/M | Severe COVID‐19 | Hypertension, diabetes, chronic renal failure | 22 days | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Axillae, groin, nostrils and ear swabs |
| 77/M | Severe COVID‐19 | Chronic obstructive pulmonary disease, heart failure, chronic renal failure | 22 days | Yes | No | Yes | No | Yes | Yes | No | Axillae, groin and nostrils |
| 64/M | Aspiration pneumonia | Tobacco and alcohol abuse, depression | 17 days | Yes | Yes | Yes | Yes | Yes | Yes | No | Axillae swab |
Abbreviations: CVC, previous or current central venous catheter; HD, previous or current haemodialysis; MV, previous or current mechanical ventilation.
First reported case.
Died before the collection of the screening body cultures.
Antifungal susceptibility testing of 45 C. auris isolates (two clinical, 18 body swabs and 25 environmental swab cultures) revealed MICs of 0.5 to 1 mg/L for AMB, 0.03 to 0.06 mg/L for voriconazole, 2–4 mg/L for fluconazole and 0.03–0.06 mg/L for anidulafungin.
Microsatellite analysis, which included 18 strains representing the different C. auris clades and six Brazilian representatives (strain L1537, patient A, CVC‐tip; strain L1686, patient B, CVC‐tip; strain L1687, bed rail room patient B; strain L1688, vital signs monitors and intravenous infusion pumps room patient B; strain L1689, digital thermometer, patient B; strain L1685, patient C, blood culture) revealed that all C. auris isolates belong to the South Asian clade (Clade I). Of note, Brazilian clinical and environmental strains were genetically closely related but belonged to three distinct STR genotypes. Strains from patient A and patient C had identical STR profiles, while the environmental strains close to patient B showed two distinct genotypes (Figure 2).
FIGURE 2.
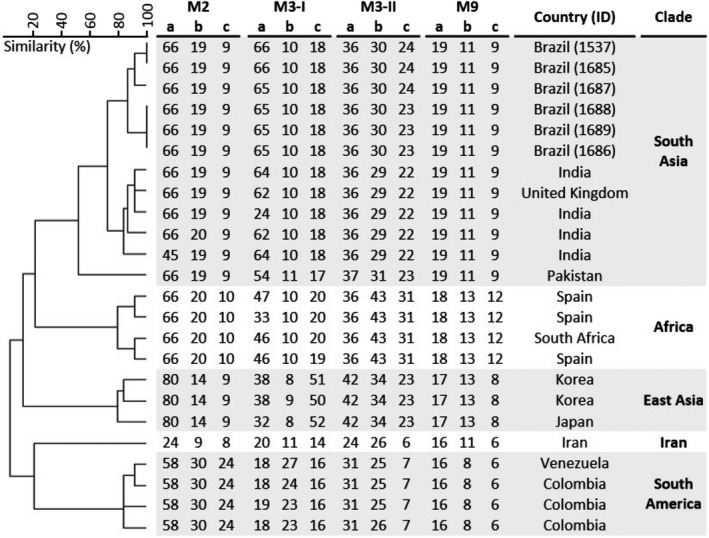
Short tandem repeat (STR) typing of Candida auris isolates from Brazil. UPGMA dendrogram of six Brazilian isolates and representative isolates from South Asian clade and other four clades is shown. Patient identification number of Brazilian isolates is indicated. The scale in the upper left corner represents similarity (%)
Investigation of risk factors associated with C. auris carriage were carried out with 9 of the 10 confirmed cases (one case was excluded because screening cultures were not collected) and 11 C. auris‐negative controls that had detailed clinical and microbiological data available. The median ages of the colonised and non‐colonised patients were 64 and 71 years, respectively. Most of the patients were male (89% colonised vs 73% non‐colonised), and 44% of the colonised and 64% of non‐colonised patients were admitted to the COVID‐19 ICU due to acute respiratory distress syndrome related to SARS‐CoV‐2 infection at admission. Overall, colonised and non‐colonised patients had similar exposure to invasive procedures, antimicrobials, antifungals and corticosteroids (Table 2). In multivariate analysis, having a positive digital thermometer culture was the only independent risk factor associated with C. auris colonisation.
TABLE 2.
Univariate and multivariate analysis of potential risk factors associated with Candida auris colonisation
| Condition | Colonised (n = 9) | Non‐colonised (n = 11) | Univariate analysis, p‐value | Multivariate analysis, p‐value (OR, CI 95%) |
|---|---|---|---|---|
| Age, median (interquartile interval – IQI) | 64 (IQI = 61–76) | 71 (IQI = 59–80) | .370 | |
| Gender, male | ||||
| Yes | 8 | 8 | .37 | |
| No | 1 | 3 | ||
| COVID‐19 ICU hospitalisation | ||||
| Yes | 4 | 7 | .653 | |
| No | 5 | 4 | ||
| Antimicrobial exposure | ||||
| Yes | 9 | 10 | 1 | |
| No | 0 | 1 | ||
| Antifungal exposure | ||||
| Yes | 5 | 4 | .653 | |
| No | 4 | 7 | ||
| Corticosteroid exposure | ||||
| Yes | 8 | 10 | 1 | |
| No | 1 | 1 | ||
| Central venous catheter | ||||
| Yes | 7 | 10 | 1 | |
| No | 2 | 1 | ||
| Urinary catheter | ||||
| Yes | 6 | 10 | .285 | .748 (0.59, 0.02–13.8) |
| No | 3 | 1 | ||
| Tracheostomy | ||||
| Yes | 4 | 6 | 1 | |
| No | 5 | 5 | ||
| Haemodialysis | ||||
| Yes | 3 | 3 | 1 | |
| No | 6 | 8 | ||
| Previous surgery | ||||
| Yes | 1 | 3 | .591 | |
| No | 8 | 8 | ||
| Colonised axillar digital thermometer | ||||
| Yes | 6 | 1 | .017 | .032 (17.1, 1.27–231.7) |
| No | 3 | 10 | ||
Values in bold were considered statistically significant.
Three patients had bloodstream infections by C. auris between December 12, 2020, and February 20, 2021. All patients died in the 30‐day follow up period after the infection. In one case, the death was attributed to the fungemia episode. Details about these three fungemia episodes are provided in Table 3.
TABLE 3.
Clinical and microbiological details of three Candida auris fungemia cases
| Condition | Episode 1 a | Episode 2 | Episode 3 |
|---|---|---|---|
| Age/sex | 74/F | 68/M | 88/F |
| Motive of Hospital Admission | Severe COVID‐19 | Upper gastrointestinal bleeding | Urinary tract infection, delirium |
| Comorbidities | Diabetes mellitus, hypertension, chronic renal failure, coronary artery disease | Diabetes mellitus, chronic renal failure, obesity | Hypertension, diabetes mellitus, dementia |
| Length of hospitalisation before fungemia | 34 days | 8 days | 11 days |
| Previous infection by MDR bacteria | Yes | Yes | No |
| Previous antifungal exposure | Yes | Yes | No |
| Invasive procedures previous fungemia | Central venous catheter, mechanical ventilation, haemodialysis | Central venous catheter, mechanical ventilation, haemodialysis | Central venous catheter, mechanical ventilation |
| Fluconazole MIC (mg/L) | 4 | 4 | 4 |
| AMB MIC (mg/L) | 1 | 1 | 1 |
| Anidulafungin MIC (mg/L) | 0.03 | 0.03 | 0.06 |
| Treatment | Anidulafungin | Anidulafungin | Anidulafungin |
| CVC removal | Yes | Yes | Yes |
| 30‐day outcome | Dead | Dead | Dead |
| Death attributed to fungemia | No | No | Yes |
Abbreviation: MDR, multidrug‐resistant bacteria, including carbapenem‐resistant Gram‐negative rods and vancomycin‐resistant enterococci.
Previously reported case. 11
4. DISCUSSION
This investigation that took place few days after the identification of the first C. auris‐colonised patient in Brazil involved a multidisciplinary team, supported by hospital infection control professionals, local and federal health agents, and researchers. The screening cultures helped to map additional C. auris‐colonised patients that were isolated and put in contact precautions. These measures, along with the substitution of disinfection products based on quaternary ammonium by sodium hypochlorite and hydrogen peroxide‐based disinfectants, 22 and replacement of all digital thermometers by infrared ones were instituted. Despite all these measures, the outbreak was not controlled, and additional screening cultures are under evaluation. Due to the rise of COVID‐19 cases in Brazil and in Salvador noticed last February and March 2021, new ICU beds were opened at the hospital, and these patients are being closely monitored for new C. auris infections by the local HICT and by the Brazilian NSSA.
In this study, most of the patients found to be colonised with C. auris had positive axillae swab cultures. The other investigated sites showed lower positivity rates and no additional cases would have been missed if they were not performed. Surprisingly, we expected more groin‐positive samples since some reports showed similar positivity rates of swabs from that body region when compared to axilla swab cultures. 16 , 20 This particular finding may be related to the high culture‐positivity rate found for the digital thermometers. Moreover, microsatellite analysis showed that clinical and thermometer strains were genetically related. Although not evaluated in this study, these colonised digital thermometers may have helped to produce a higher fungal burden at the axillae of these patients. Indeed, axillary temperature monitoring with reusable probes was an independent risk factor for C. auris colonisation in a UK outbreak. 17 Therefore, hospital epidemiologists and infection control professionals have to be aware that axillary temperature monitoring with insufficiently disinfected reusable probes or digital thermometers may facilitate the dissemination of C. auris in hospital settings. Before recognition of this outbreak, the digital thermometers were disinfected with quaternary ammonium compounds, which are most likely insufficient for the elimination of C. auris biofilms. 22 , 23 More stringent disinfection methods may be an alternative for the thermometer's replacement. 24 In addition to positive cultures of the digital thermometers, more than 10% of the bed railing and tray table samples were positive for C. auris, confirming that C. auris is able to persist on different inanimate surfaces in the patients’ vicinities. Our findings are similar of those found in New York state hospitals, where 12 to 21% of the environmental screening cultures were positive for C. auris. 25 The widespread presence of C. auris in the patient's vicinities, including highly manipulated regions, facilitates the occurrence of missing hand disinfection opportunities and contributes to the horizontal transmission of this pathogen. In the context of the COVID‐19 pandemic, one may expect an increase in C. auris infections around the world. Overwhelmed ICUs with a scenario of mechanically ventilated patients and with other invasive procedures, high antimicrobial and antifungal selective pressure, and reuse of personal protective equipment due to shortages, may lead to the selection and horizontal transmission of C. auris. In USA, India, Mexico, Italy, Lebanon and now in Brazil, outbreaks of C. auris colonisation/infection are being reported in COVID‐19 ICUs. 7 , 8 , 9 , 26 , 27
Outbreaks of C. auris are usually associated with fluconazole resistance, with at least more than 10% of the isolates considered AMB resistant. 15 , 17 , 28 , 29 In contrast, all forty‐five strains we analysed had low AMB, fluconazole and anidulafungin MICs. We previously reported that the first strain had wildtype ERG11 and FKS1 DNA sequences. 11 Although not evaluated in this study, we assume that all isolates lack the ERG11 and FKS1 hotspot mutations due to the persistent low azole and anidulafungin MICs. These peculiar findings support the hypothesis that C. auris was recently introduced into the Brazilian hospital environment and antifungal exposure over that short period of time was not sufficient to induce resistance.
Despite the short period between the first diagnosis and the cross‐sectional interventional investigation, genetic diversity was already noticed among the Brazilian C. auris isolates. Similarly, the investigation carried out in the United Kingdom revealed that 22% of the patients had mixed C. auris genotypes colonisation/infection at their first positive culture result. 17 These findings should warrant clinical microbiologists that mixed populations may be common among C. auris‐positive cultures, and antifungal susceptibility testing should be carried from different colony morphologies or subpopulations. Despite the good in vitro antifungal activity, 1 log MIC value differences were noted for some subpopulations from the same isolate of our cohort.
Previous empiric use of antifungals, including fluconazole, exposure to corticosteroids, tracheostomy and haemodialysis, were highly prevalent among C. auris‐colonised and non‐colonised patients. The only independent risk factor for C. auris colonisation in this study was the axillary temperature monitoring with contaminated reusable digital thermometers. These findings may be related to the use of quaternary ammonium disinfection, 22 and most patients hospitalised during the outbreak were admitted due to severe COVID‐19 and had a similar clinical course.
Finally, as previously reported, bloodstream infections by C. auris occurred in patients with severe baseline underlying diseases after long periods of hospital admission and exposure to antimicrobials, central venous catheterisation and mechanical ventilation. 30 , 31 , 32 , 33 In this scenario, the high crude mortality is multifactorial and not only attributed to C. auris infection. 34
In conclusion, it is intriguing that the Brazilian C. auris cluster is completely different from the predominant clusters already described in Venezuela, Colombia and Panama. 18 , 35 , 36 The emergence of a multi‐susceptible strain of C. auris in Salvador, Brazil, may be related to local C. auris clade I closely related genotypes. The outbreak related to C. auris isolates with low MICs is unusual, but antifungal susceptibility vigilance is required due to the high potential of antifungal resistance development of this species. Contaminated axillary monitoring thermometers may facilitate the dissemination of C. auris reinforcing the concept that these reusable devices should be carefully cleaned with an effective disinfectant or replaced by other temperature monitoring methods, mainly in locations where C. auris is emerging.
CONFLICT OF INTEREST
ALC received educational grants from Eurofarma, Pfizer, Gilead Sciences, United Medical (Brazil), Knight, TEVA. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
Joao Nobrega de Almeida Júnior: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Investigation (lead); Methodology (lead); Project administration (supporting); Validation (supporting); Writing‐original draft (lead); Writing‐review & editing (equal). Igor Brandão: Data curation (equal); Investigation (equal); Resources (equal); Supervision (equal); Writing‐original draft (supporting); Writing‐review & editing (equal). Elaine Cristina Francisco: Data curation (lead); Formal analysis (equal); Methodology (equal); Project administration (equal); Supervision (equal); Writing‐original draft (supporting). Silvio Luis Rodrigues: Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Validation (equal); Writing‐review & editing (equal). Patrícia de Oliveira Dias: Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (supporting); Project administration (supporting); Validation (equal). Felicidade Pereira: Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Writing‐review & editing (equal). Fabio Ferreira: Investigation (equal); Methodology (equal); Supervision (equal); Validation (equal); Writing‐review & editing (equal). Thaisse Andrade: Data curation (equal); Formal analysis (equal); Investigation (equal); Supervision (equal); Writing‐review & editing (equal). Magda Miranda: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Project administration (equal); Resources (equal); Supervision (equal); Writing‐review & editing (equal). Regiane Jordão: Formal analysis (equal); Investigation (equal); Project administration (equal); Resources (equal); Supervision (equal); Writing‐review & editing (equal). Jacques F. Meis: Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Resources (equal); Validation (equal); Writing‐review & editing (equal). Arnaldo Lopes Colombo: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Funding acquisition (lead); Investigation (equal); Methodology (equal); Project administration (lead); Resources (lead); Supervision (equal); Validation (lead); Writing‐original draft (lead); Writing‐review & editing (equal). Larissa Molina: Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Writing‐original draft (supporting). Soraia Lima: Data curation (supporting); Formal analysis (supporting); Methodology (supporting); Project administration (supporting); Supervision (supporting); Validation (supporting). Ricardo Lima: Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Validation (supporting). Ismaiane Miranda: Data curation (supporting); Investigation (supporting); Methodology (supporting). Tamara Jesus: Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Methodology (supporting); Resources (supporting). Danniely Silva: Data curation (supporting); Investigation (supporting); Methodology (supporting); Project administration (supporting); Validation (supporting). Lilian Moura: Data curation (supporting); Investigation (supporting); Methodology (supporting); Project administration (supporting). Laíse Ribeiro: Data curation (supporting); Investigation (supporting); Methodology (supporting); Project administration (supporting). Antonio Carlos Bandeira: Formal analysis (supporting); Investigation (supporting); Methodology (supporting); Project administration (supporting); Resources (supporting). Talita Urpia: Data curation (supporting); Investigation (supporting); Methodology (supporting); Project administration (supporting). Mara Gonçalves: Formal analysis (supporting); Investigation (supporting); Project administration (supporting); Resources (supporting); Supervision (supporting); Validation (supporting). Theun de Groot: Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Methodology (supporting).
Supporting information
Table S1
ACKNOWLEDGMENTS
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP): 2017/02203‐7. JNAJ has received a scholarship grant (FAPESP 2018/19347‐4). We would like to thank Adriana L. Motta from Hospital das Clínicas de São Paulo for its technical assistance.
APPENDIX 1.
Members of the Candida auris Brazilian Study Group
Larissa M. Favarello (Disciplina de Infectologia; Escola Paulista de Medicina, Universidade Federal de São Paulo; São Paulo, Brazil), Soraia L. Lima (Disciplina de Infectologia; Escola Paulista de Medicina, Universidade Federal de São Paulo; São Paulo, Brazil), Ricardo Lima (Disciplina de Infectologia; Escola Paulista de Medicina, Universidade Federal de São Paulo; São Paulo, Brazil), Ismaiane Oliveira Miranda (Central Laboratory Division, Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil), Tamara L. de Jesus Lopes (Central Laboratory Division, Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil), Danniely C. Soares da Silva (Comissão de Controle de Infecção Hospitalar, Hospital de Bahia, Salvador, Brazil), Lilian Nobre de Moura (Comissão de Controle de Infecção Hospitalar, Hospital de Bahia, Salvador, Brazil), Laísse C. Ribeiro (Laboratório Central de Saúde Pública Professor Gonçalo Muniz, Salvador, Brazil), Antonio Carlos de Albuquerque Bandeira (Superintendência de Vigilância e Proteção da Saúde, Secretaria de Saúde do Estado da Bahia, Salvador, Brazil), Talita Moreira Urpia (Superintendência de Vigilância e Proteção da Saúde, Secretaria de Saúde do Estado da Bahia, Salvador, Brazil), Mara Rubia Gonçalves (Brazilian Health Regulatory Agency, Ministério da Saúde, Brasília, Brazil), Theun de Groot (Department of Medical Microbiology and Infectious Diseases, ECMM Center of Excellence for Medical Mycology, 6532 SZ Nijmegen, The Netherlands).
Nobrega de Almeida J Jr, Brandão IB, Francisco EC, et al; The Candida auris Brazilian Study Group . Axillary Digital Thermometers uplifted a multidrug‐susceptible Candida auris outbreak among COVID‐19 patients in Brazil. Mycoses. 2021;64:1062–1072. 10.1111/myc.13320
Contributor Information
Arnaldo L. Colombo, Email: arnaldolcolombo@gmail.com.
The Candida auris Brazilian Study Group:
Larissa M Favarello, Soraia L. Lima, Ricardo Lima, Ismaiane Oliveira Miranda, Tamara L. de Jesus Lopes, Danniely C. Soares da Silva, Lilian Nobre de Moura, Laísse C. Ribeiro, Antonio Carlos de Albuquerque Bandeira, Talita Moreira Urpia, Mara Rubia Gonçalves, and Theun de Groot
DATA AVAILABILITY STATEMENT
All sequences were deposited and are available at GenBank (supplementary material, https://www.ncbi.nlm.nih.gov/genbank/).
REFERENCES
- 1. Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug‐resistant Candida auris on 3 continents confirmed by whole‐genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134‐140. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meis JF, Chowdhary A Candida auris: a global fungal public health threat. Lancet Infect Dis. 2018;18(12):1298‐1299. 10.1016/S1473-3099(18)30609-1 [DOI] [PubMed] [Google Scholar]
- 3. Lone SA, Ahmad A Candida auris‐the growing menace to global health. Mycoses. 2019;62(8):620‐637. 10.1111/myc.12904 [DOI] [PubMed] [Google Scholar]
- 4. Govender NP, Magobo RE, Mpembe R, et al. Candida auris in South Africa, 2012–2016. Emerg Infect Dis. 2018;24(11):2036‐2040. 10.3201/eid2411.180368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chowdhary A, Anil Kumar V, Sharma C, et al. Multidrug‐resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis. 2014;33(6):919‐926. 10.1007/s10096-013-2027-1 [DOI] [PubMed] [Google Scholar]
- 6. Shastri PS, Shankarnarayan SA, Oberoi J, et al. Candida auris candidaemia in an intensive care unit ‐ prospective observational study to evaluate epidemiology, risk factors, and outcome. J Crit Care. 2020;57:42‐48. 10.1016/j.jcrc.2020.01.004 [DOI] [PubMed] [Google Scholar]
- 7. Prestel C, Anderson E, Forsberg K, et al. Candida auris outbreak in a COVID‐19 specialty care unit ‐ Florida, July‐August 2020. MMWR Morb Mortal Wkly Rep. 2021;70(2):56‐57. 10.15585/mmwr.mm7002e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Villanueva‐Lozano H, Treviño‐Rangel RDJ, González GM, et al. Outbreak of Candida auris infection in a COVID‐19 hospital in Mexico. Clin Microbiol Infect. 2021;27(5):813‐816. 10.1016/j.cmi.2020.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Magnasco L, Mikulska M, Giacobbe DR, et al. Spread of carbapenem‐resistant gram‐negatives and Candida auris during the COVID‐19 pandemic in critically Ill patients: one step back in antimicrobial stewardship? Microorganisms. 2021;9(1):95. 10.3390/microorganisms9010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pasqualotto AC, Sukiennik TCT, Meis JF. Brazil is so far free from Candida auris. Are we missing something? Braz J Infect Dis. 2019;23(3):149‐150. 10.1016/j.bjid.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Almeida JN, Francisco EC, Hagen F, et al. Emergence of Candida auris in Brazil in a COVID‐19 intensive care unit. J Fungi. 2021;7(3):220. 10.3390/jof7030220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caceres DH, Forsberg K, Welsh RM, et al. Candida auris: a review of recommendations for detection and control in healthcare settings. J Fungi. 2019;5(4):111. 10.3390/jof5040111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kenters N, Kiernan M, Chowdhary A, et al. Control of Candida auris in healthcare institutions: Outcome of an International Society for Antimicrobial Chemotherapy expert meeting. Int J Antimicrob Agents. 2019;54(4):400‐406. 10.1016/j.ijantimicag.2019.08.013 [DOI] [PubMed] [Google Scholar]
- 14. Gerência de Vigilância e Monitoramento em Serviços de Saúde , Gerência Geral de Tecnologia em Serviços de Saúde , Agência Nacional de Vigilância Sanitária . COMUNICADO DE RISCONo 01/2017–GVIMS/GGTES/ANVISA. Relatos de surtos de Candida auris em serviços de saúde da América Latina; 2017.
- 15. Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:35. 10.1186/s13756-016-0132-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruiz‐Gaitán A, Moret AM, Tasias‐Pitarch M, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61(7):498‐505. 10.1111/myc.12781 [DOI] [PubMed] [Google Scholar]
- 17. Eyre DW, Sheppard AE, Madder H, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379(14):1322‐1331. 10.1056/NEJMoa1714373 [DOI] [PubMed] [Google Scholar]
- 18. Escandón P, Chow NA, Caceres DH, et al. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in Amphotericin B resistance. Clin Infect Dis. 2019;68(1):15‐21. 10.1093/cid/ciy411 [DOI] [PubMed] [Google Scholar]
- 19. Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109(16):6241‐6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clinical and Laboratory Standards Institute . Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 4th ed. CLSI standard M27. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 21. de Groot T, Puts Y, Berrio I, et al. Development of Candida auris short tandem repeat typing and its application to a global collection of isolates. MBio. 2020;11(1):e02971‐19. 10.1128/mBio.02971-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sexton DJ, Welsh RM, Bentz ML, et al. Evaluation of nine surface disinfectants against Candida auris using a quantitative disk carrier method: EPA SOP‐MB‐35. Infect Control Hosp Epidemiol. 2020;41(10):1219‐1221. 10.1017/ice.2020.278 [DOI] [PubMed] [Google Scholar]
- 23. Ku TSN, Walraven CJ, Lee SA Candida auris: disinfectants and implications for infection control. Front Microbiol. 2018;9:726. 10.3389/fmicb.2018.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meis JF, Voss A Candida auris in an intensive care setting. N Engl J Med. 2019;380(9):890‐891. 10.1056/NEJMc1900112 [DOI] [PubMed] [Google Scholar]
- 25. Adams E, Quinn M, Tsay S, et al. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis. 2018;24(10):1816‐1824. 10.3201/eid2410.180649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chowdhary A, Tarai B, Singh A, et al. Multidrug‐resistant candida auris infections in critically Ill coronavirus disease patients, India, April‐July 2020. Emerg Infect Dis. 2020;26(11):2694‐2696. 10.3201/eid2611.203504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allaw F, Kara Zahreddine N, Ibrahim A, et al. First Candida auris outbreak during a COVID‐19 pandemic in a Tertiary‐Care Center in Lebanon. Pathogens. 2021;10(2):157. 10.3390/pathogens10020157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Armstrong PA, Rivera SM, Escandon P, et al. Hospital‐associated multicenter outbreak of emerging fungus Candida auris, Colombia, 2016. Emerg Infect Dis. 2019;25(7):1339‐1346. 10.3201/eid2507.180491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alfouzan W, Ahmad S, Dhar R, et al. Molecular epidemiology of Candida auris outbreak in a major secondary‐care hospital in Kuwait. J Fungi. 2020;6(4):307. 10.3390/jof6040307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Di Pilato V, Codda G, Ball L, et al. Molecular epidemiological investigation of a nosocomial cluster of C auris: evidence of recent emergence in Italy and ease of transmission during the COVID‐19 pandemic. J Fungi. 2021;7(2):140. 10.3390/jof7020140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Almaghrabi RS, Albalawi R, Mutabagani M, et al. Molecular characterisation and clinical outcomes of Candida auris infection: single‐centre experience in Saudi Arabia. Mycoses. 2020;63(5):452‐460. 10.1111/myc.13065 [DOI] [PubMed] [Google Scholar]
- 32. Mohsin J, Weerakoon S, Ahmed S, et al. A cluster of Candida auris blood stream infections in a Tertiary Care Hospital in Oman from 2016 to 2019. Antibiotics. 2020;9(10):638. 10.3390/antibiotics9100638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahmad S, Khan Z, Al‐Sweih N, et al. Candida auris in various hospitals across Kuwait and their susceptibility and molecular basis of resistance to antifungal drugs. Mycoses. 2020;63(1):104‐112. 10.1111/myc.13022 [DOI] [PubMed] [Google Scholar]
- 34. Chakrabarti A, Singh S. Multidrug‐resistant Candida auris: an epidemiological review. Expert Rev Anti Infect Ther. 2020;18(6):551‐562. 10.1080/14787210.2020.1750368 [DOI] [PubMed] [Google Scholar]
- 35. Calvo B, Melo ASA, Perozo‐Mena A, et al. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect. 2016;73(4):369‐374. 10.1016/j.jinf.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 36. Araúz AB, Caceres DH, Santiago E, et al. Isolation of Candida auris from 9 patients in Central America: importance of accurate diagnosis and susceptibility testing. Mycoses. 2018;61(1):44‐47. 10.1111/myc.12709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
All sequences were deposited and are available at GenBank (supplementary material, https://www.ncbi.nlm.nih.gov/genbank/).


