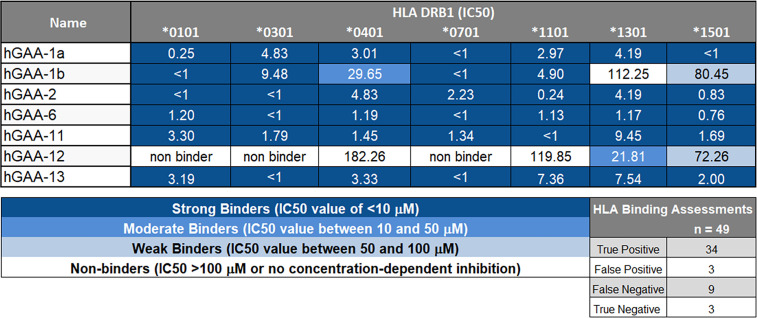
Figure 4.
GAA-peptides bind to HLA DR1 as predicted. Selected GAA peptides were evaluated for HLA DRB1 binding in vitro and IC50 values were calculated. hGAA-1a, hGAA-1b, hGAA-2, hGAA-6, hGAA-11 and hGAA-13 FV621 peptides bound with the multiple alleles tested (DRB1*0101, *0301, *0401, *0701, *1101, *1301 and *1501) whereas hGAA-12 was predicted to be more HLA-restricted and consequently had limited binding to HLA. A seven-point competition assay using a validated control peptide was performed; color coding reflects binding affinity IC50 was determined by interpolation. Using a standard Z-score threshold of 1.64 (top 5%), overall positive predictive value for EpiMatrix predictions was 92%, with sensitivity of 79%. False negatives are not uncommon when testing peptides containing significant predictions for several alleles (EpiBars), as many contain “near-miss” Z-scores in the top 10% of predicted peptides. Note that peptide GAA-12 and 13 were not selected for promiscuity: they were designed for individualized testing in a specific patient for which both the mutation and the HLA DRB1 allele restriction concurred.