Figure 6.
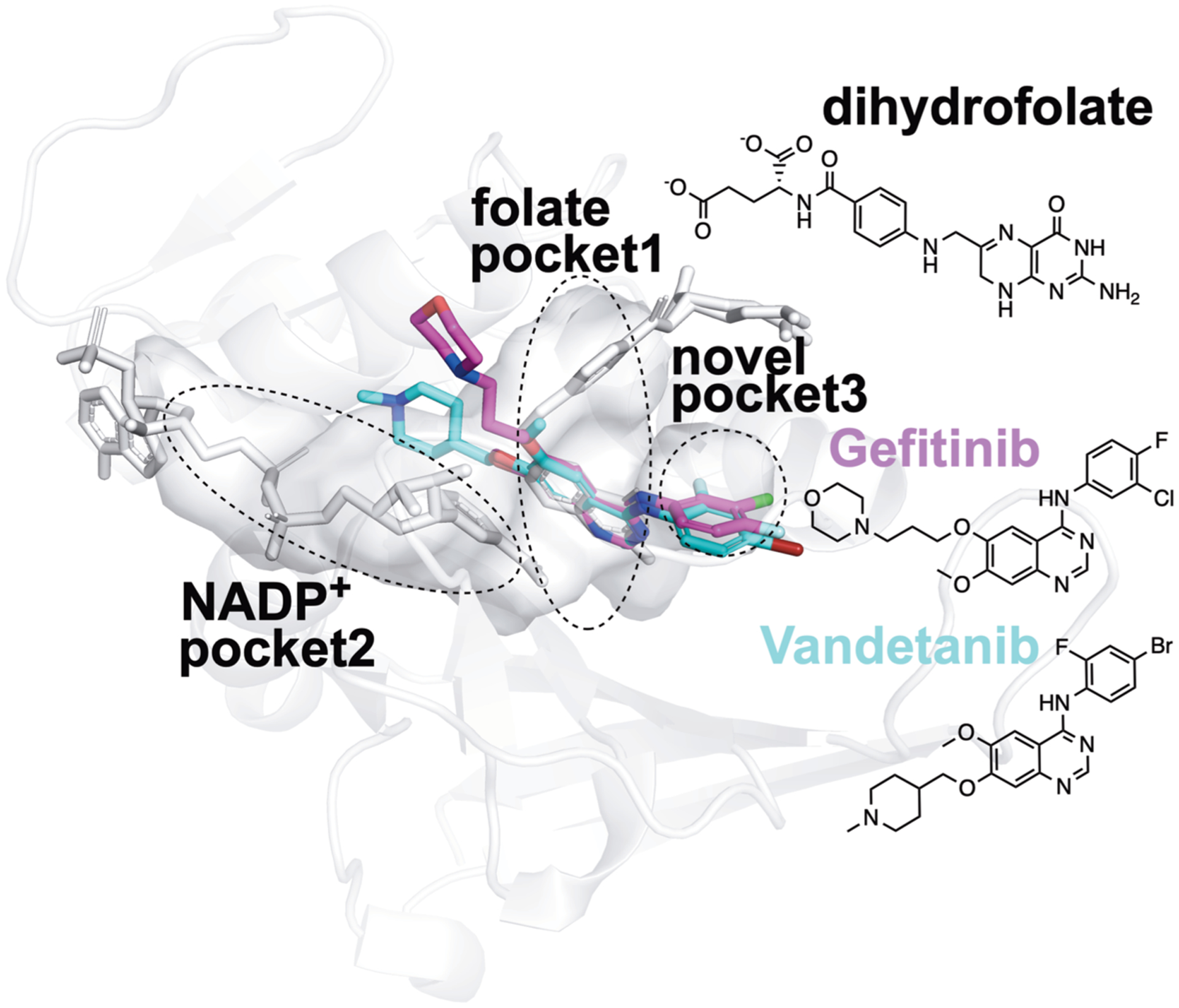
Proposed structural basis of the new true binders of E. coli DHFR identified by FRAGSITE. Using the PyMOL Molecular Graphics System Version 2.3.2 (Schrödinger, LLC), the PDB ligand model structures of NSC715055 (Gefitinib, PDB ligand code IRE, shown as purple sticks) and NSC760766 (Vandetanib, PDB ligand code ZD6, shown as cyan sticks) were manually and rigidly docked into ligand binding pockets in the E. coli DHFR:folate:NADP+ ternary complex crystal structure (PDB entry 4PSY,65 pockets shown as the van der Waals surface in white color) based on spatial alignment of the bicyclic core of the new true binders identified by FRAGSITE to the pterin moiety of the folate ligand in the crystal structure. Based on the structural alignment and the conceivably rotatable O and N-linkages extended from the bicyclic core of these new ligands, their proposed binding modes in E. coli DHFR may result in occupying not only the dihydrofolate pocket but also NADPH and a third new pocket. The protein secondary structures are shown as white ribbons, and the original crystallographically identified ligands (NADP+ and dihydrofolate) in PDB entry 4PSY65 are shown as white sticks. The chemical structures of Gefitinib, Vandetanib, and dihydrofolate are also shown for comparison.
