Figure 3. Role of myeloid cell tropism in RhCMV vector CD8+ T cell response programming.
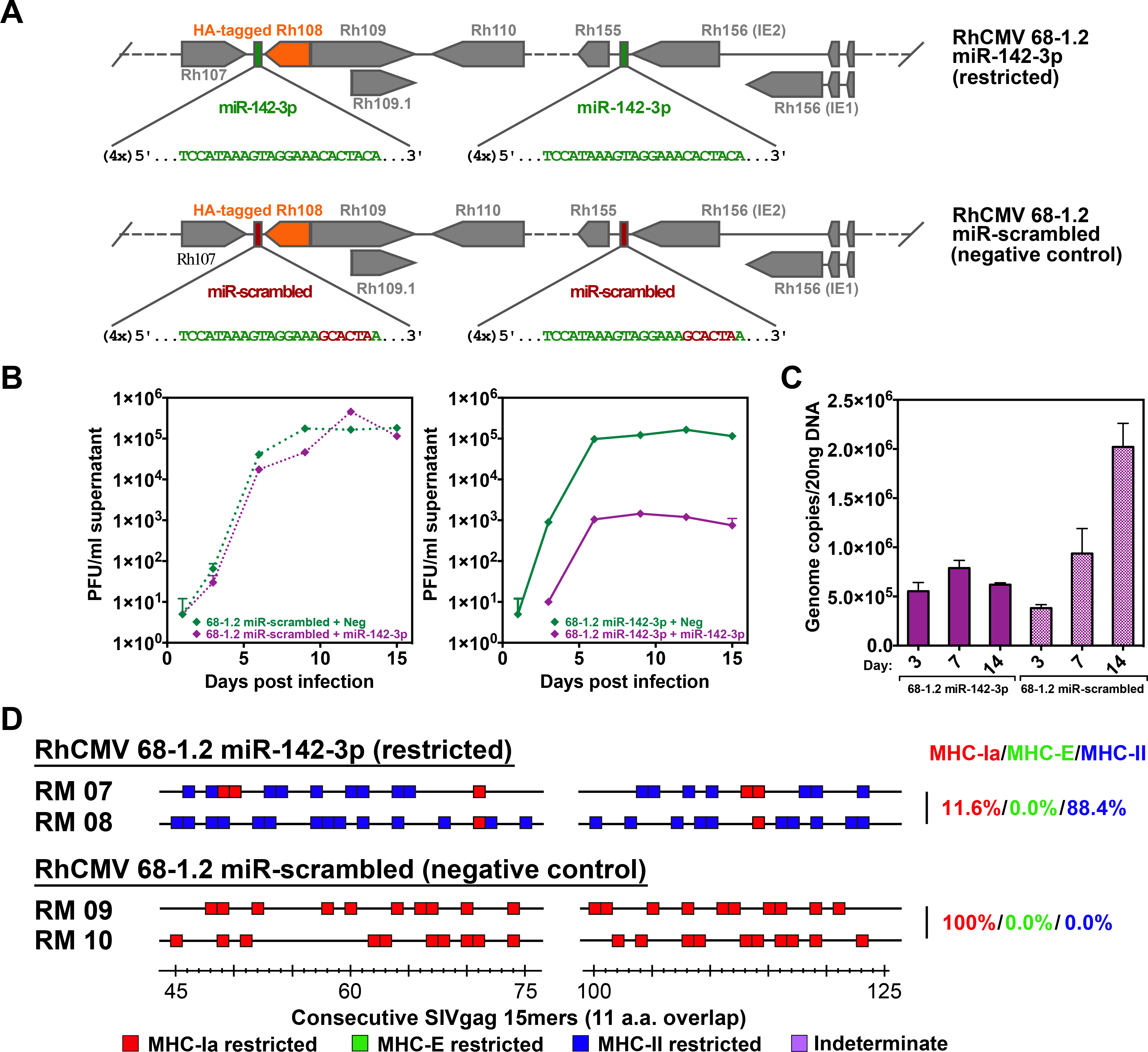
(A) Schematic of the miR-142–3p target sites inserted downstream of both Rh156 and Rh108 (homologs of HCMV IE2 and UL79), which are essential for viral replication. Green sequences represent insertion of four miR-142–3p recognition sites. Red sequences indicate scrambled nucleotides of the miR-142–3p sequence used to create a control vector. Orange boxes represent the Rh108 ORF with a hemagglutinin (HA) epitope-tag. (B) Growth analysis of the 68–1.2 control (scrambled sequence) RhCMV vector (left) vs. 68–1.2 miR-142–3p RhCMV vector (right) in the presence or absence of miR-142–3p expression. Primary rhesus fibroblasts were transfected with negative control or miR-142–3p mimic and infected 24 hours later with the 68–1.2 miR-142–3p or control RhCMV vectors at an MOI of 0.01. Cell supernatants were harvested at the indicated timepoints and titered on primary rhesus fibroblasts. Results are representative of 2 independent experiments. (C) Analysis of 68–1.2 miR-142–3p vs control RhCMV vector replication in primary macrophages. Macrophages were differentiated in vitro from peripheral blood of 3 RMs and infected with 68–1.2 miR-142–3p or scrambled RhCMV at MOI = 5. At the indicated times post-infection viral DNA was isolated and total DNA copies were determined using qPCR (mean + SEM of 3 independent experiments shown). (D) Representative MHC restriction analysis of SIVgag-specific CD8+ T cell responses elicited by 68–1.2 miR-142–3p vs. scrambled RhCMV/SIVgag vectors, as described in Fig. 1. Overall epitope analysis results are shown in table S1.
