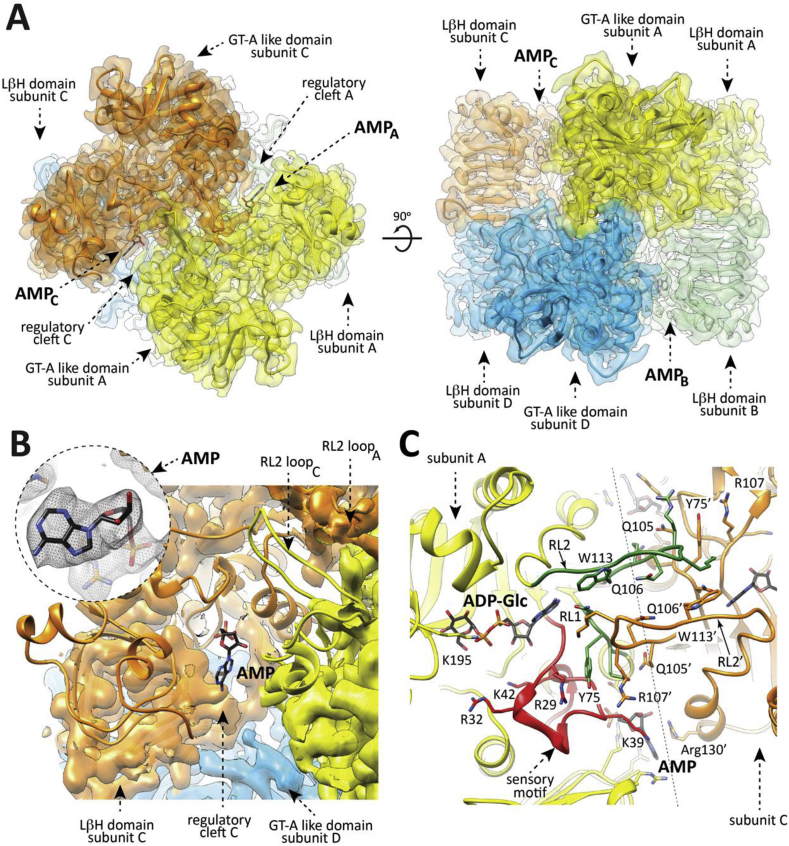
Fig. 3.
Overall structure of the EcAGPase-AMP complex as visualized by cryoEM. (A) The overall view of the electron density cryoEM map of the EcAGPase-AMPD2 complex, colored according to the four protomers (orange, yellow, blue and green) of the homotetramer. (B) A selected region corresponding to the neighborhood of the AMP binding site is masked to reveal the atomic model built into the electron density, with the protein shown as ribbons and the ligand AMP in sticks representation. Close-up of the EcAGPase-AMPD2 regulatory site showing the location and the electron density cryoEM map of the activator AMP. (C) Cartoon representation of the interface between the reference protomer (yellow) and the neighbor subunit (orange) of the EcAGPase-AMPD2 reconstruction. Note that the RL2 loop conformation, as observed in the EcAGPase-AMPD2 reconstruction, is oriented towards the neighbor active site.