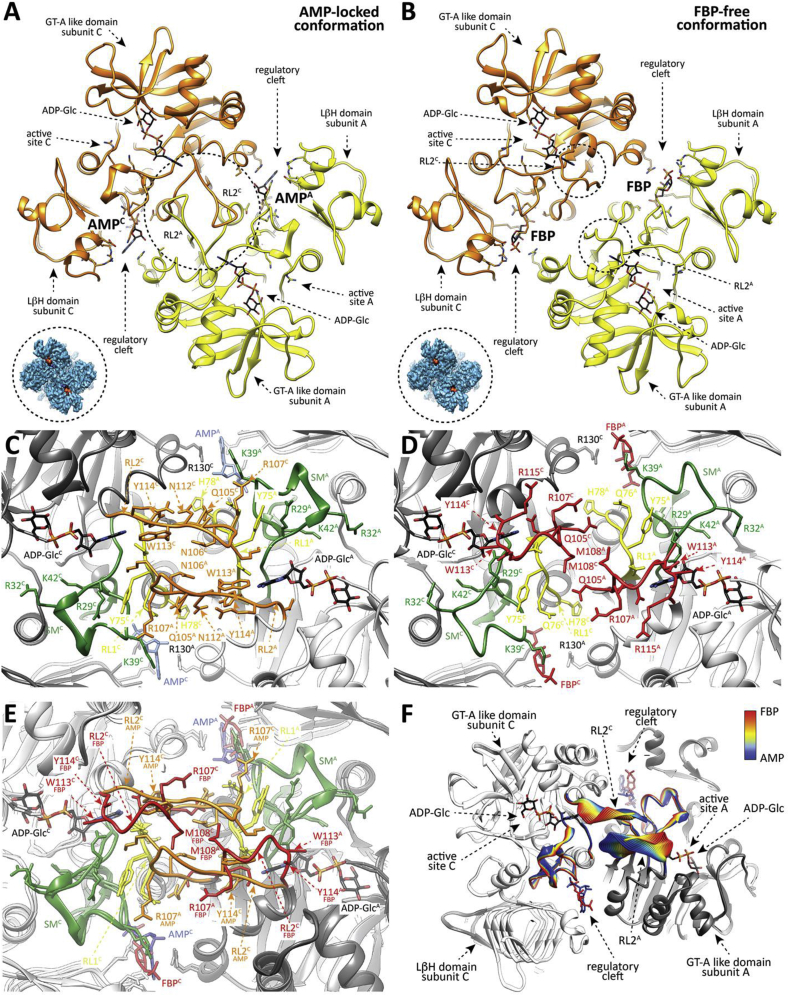
Fig. 5.
The conformation of the RL2 loop as observed in the EcAGPase-FBPD2and EcAGPase-AMPD2complexes. (A) View of two neighboring protomers of different dimers (yellow and orange) in the EcAGPase-FBPD2 complex. FBP is observed in the regulatory cleft, whereas ADP-Glc is shown in the active site as a reference. The “free” conformation of the RL2 loops is indicated. A caption of the EcAGPase-FBPD2 complex map (blue) is shown with ADP-Glc placed in the active site. (B) As in panel (A), two neighboring protomers of different dimers in the EcAGPase-AMPD2 complex. AMP is observed in the regulatory cleft, whereas ADP-Glc is shown in the active site as a reference. The “locked” conformation of the RL2 loops is indicated. A caption of the EcAGPase-AMPD2 complex map (blue) is shown with ADP-Glc to observe map differences of the active shape compared with the EcAGPase-FBPD2 complex. (C) Closed view of the SM, RL1 and RL2 loops as observed in the EcAGPase-AMPD2 complex. (D) Closed view of the SM, RL1 and RL2 loops as observed in the EcAGPase-FBPD2 complex. (E) Structural comparison of the RL1 and RL2 loops as observed in the EcAGPase-FBPD2 and EcAGPase-AMPD2 complexes. (F) Cartoon representation showing the transition of the SM, RL1 and RL2 loops conformation as observed by cryoEM. Different positions of the main chain are colored according to the color scheme shown in the bar (FBP complex red, AMP complex blue).