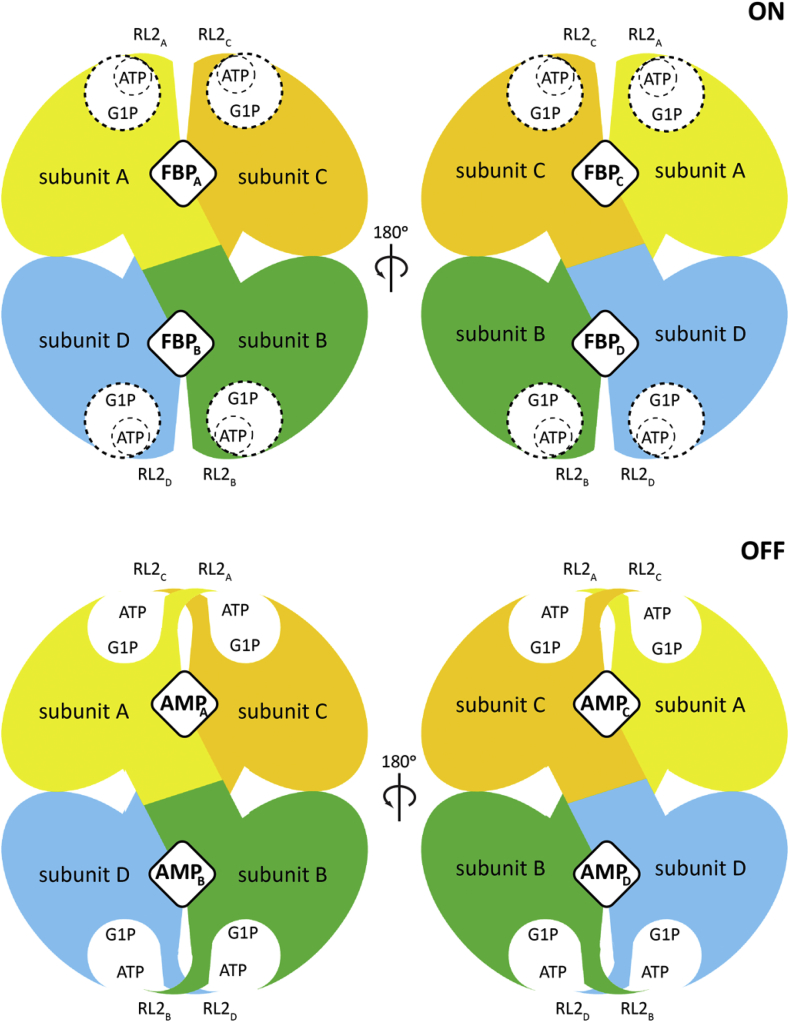
Fig. 8.
A molecular model for the allosteric regulation of EcAGPase. The EcAGPase tetramer is displayed in overlapping purple spheres, where the dotted lines are inter-protomer interphases. The active site is displayed as an open circle containing the substrates ATP and G1P. The allosteric clefts are indicated with rhombi at the interphases, that can alternatively be occupied by FBP or AMP, leading to the ON or OFF states, respectively. In the enzyme ON-state, the active site's RL2 loops (depicted in colors) are in a putative conformation allowing the interaction with ATP. In contrast, in the OFF inhibited AMP-state, the RL2 loops from neighbor protomers are engaged in the “locked” state, sequestering the loops for the interaction with ATP.