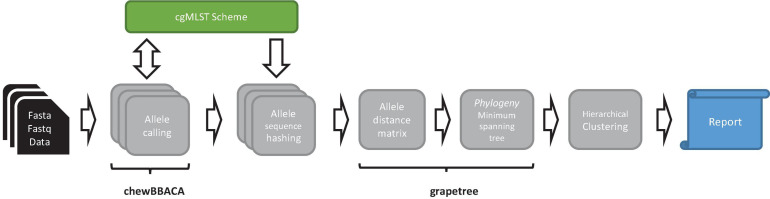
FIGURE 1.
Schematic representation of the chewieSnake workflow. The workflow starts from a set of assemblies (fasta data) or a set of raw reads (fastq data), which are then assembled into fasta files. On these, cgMLST allele calling is performed using the tool chewBBACA, utilizing a designated cgMLST scheme, to which newly found alleles are added in the process. The resulting allele profiles are converted to hashed allele profiles. All profiles are subsequently combined; and an allele distance matrix and a minimum-spanning tree phylogeny are computed with the tool GrapeTree. Then, cluster types are generated by hierarchical clustering at different allele distance thresholds. Finally, the results are summarized in an interactive HTML report.