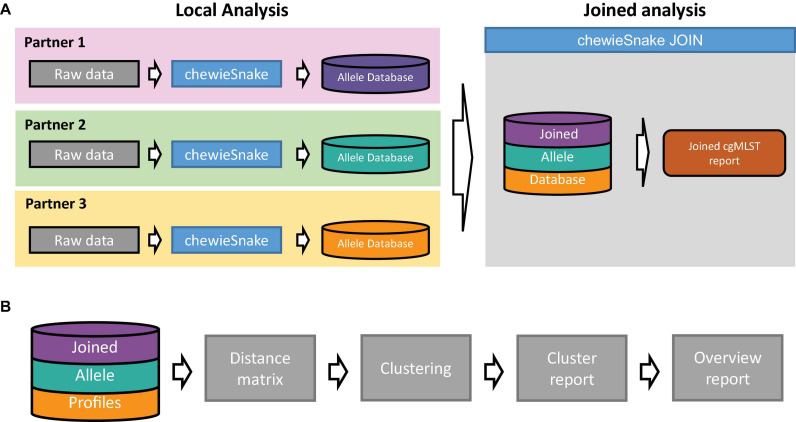
FIGURE 2.
Schematic representation of the chewieSnake_join workflow. (A) A set of laboratories locally employ the chewieSnake workflow to process incoming raw sequencing data. The resulting allele profiles are stored in individual allele databases, which are subsequently joined with the chewieSnake_join workflow. In addition, the chewieSnake_join workflow computes a joined cgMLST report. (B) Detailed description of the chewieSnake_join workflow. From the joined allele profiles, the allele distance matrix is computed, followed by cluster typing of the entire dataset. Detailed reports for each cluster as well as an overall report summarize the results in an interactive HTML format.