Figure 1.
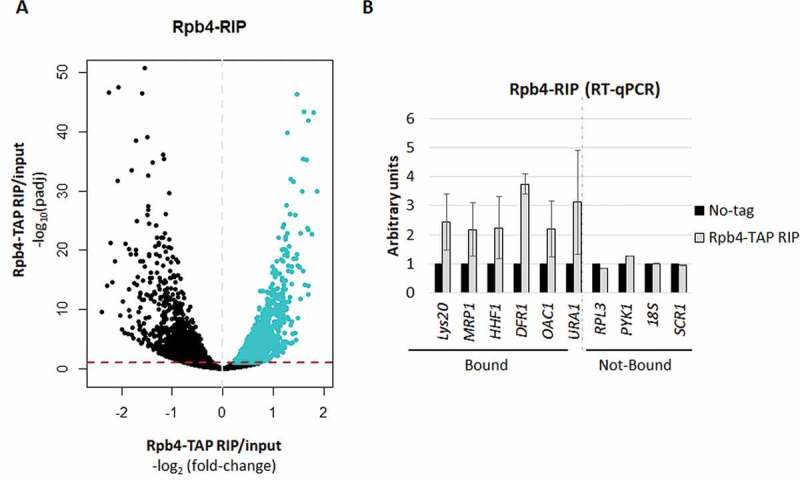
RNA immunoprecipitation (RIP) for Rpb4. (A) Volcano-plot generated to identify the significantly bound mRNAs by Rpb4 (blue), calculated by the DESeq2 software (RIP vs. input) [52]. A specific cut-off was applied to define the Rpb4-bounded transcripts consisting of Log2 Fold-Change of RIP vs. input higher than 0 (dashed grey line) and padj<0.1 (dashed red line). (B) RT-qPCR to analyse several Rpb4-bound and unbound mRNAs from the RIP experiments corresponding to the Rpb4-TAP and the no-TAP tag control strains. Data are shown as the average and standard deviation (SD) of at least three independent experiments (RIP vs. input). rRNA 18S was used as a normalizer
