Figure 3.
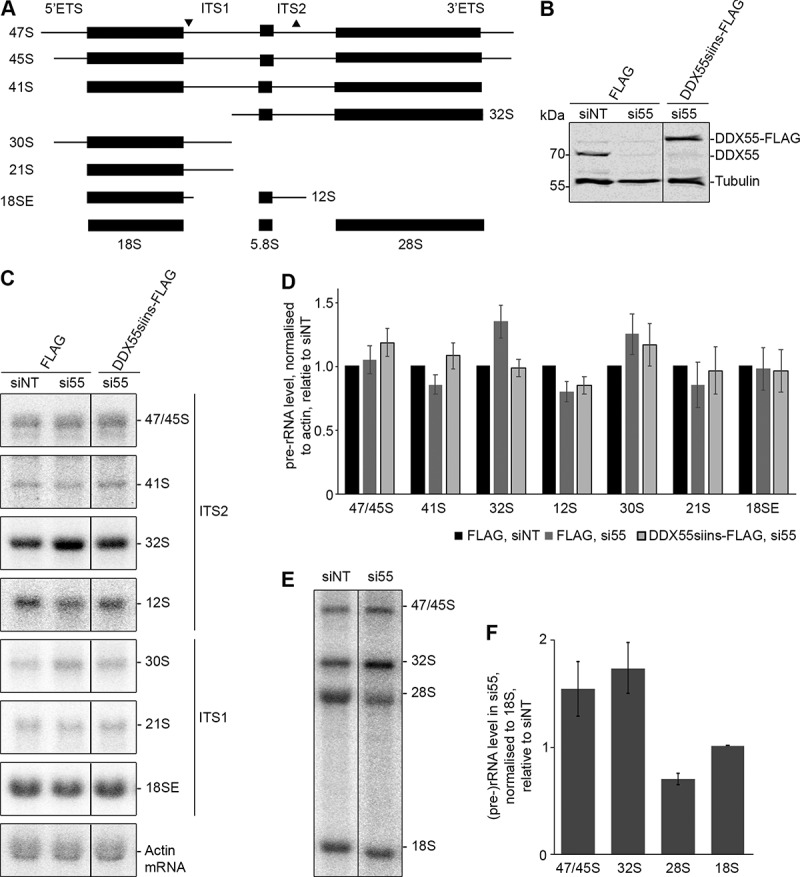
Pre-rRNA processing in cells depleted of DDX55. (A) Simplified processing scheme showing the major pre-rRNA intermediates present in human cells. Mature rRNA sequences are shown as black rectangles, and internal and external transcribed spacers (ITS and ETS respectively) are represented by black lines. The hybridization positions of probes used for northern blotting are indicated by triangles on the initial 47S pre-rRNA transcript. (B) HEK293 cells expressing the Flag tag or siRNA-insensitive, Flag-tagged DDX55 (DDX55siins-Flag) were treated with siRNAs targeting the firefly luciferase (siNT) or DDX55 (si55). Cells were harvested 90 h after transfection, and proteins were separated by SDS-PAGE and analysed by western blot using antibodies against DDX55 (α-DDX55) or, as a loading control tubulin (α-tubulin). (C) Total RNA prepared from siRNA-treated cells as in (B) was separated by denaturing agarose gel electrophoresis and transferred to a nylon membrane. Pre-rRNA species and the actin mRNA were detected by northern blotting using the probes indicated to the right of the panel. (D) The levels of pre-rRNA species in three independent experiments were quantified, normalized according to the actin mRNA, and are presented relative to the Flag, siNT sample as mean ± standard error. (E) HeLa cells treated with siNT or siDDX55 were subjected to pulse-chase metabolic labelling. Total RNAs were separated by denaturing agarose gel electrophoresis and transferred to a nylon membrane. Abundant labelled RNAs were detected using a phosphorimager. (F) The levels of (pre-)rRNA species in three independent pulse-chase experiments were quantified, normalized according to the 18S rRNA, and are presented relative to the siNT sample as mean ± standard error
