Figure 6.
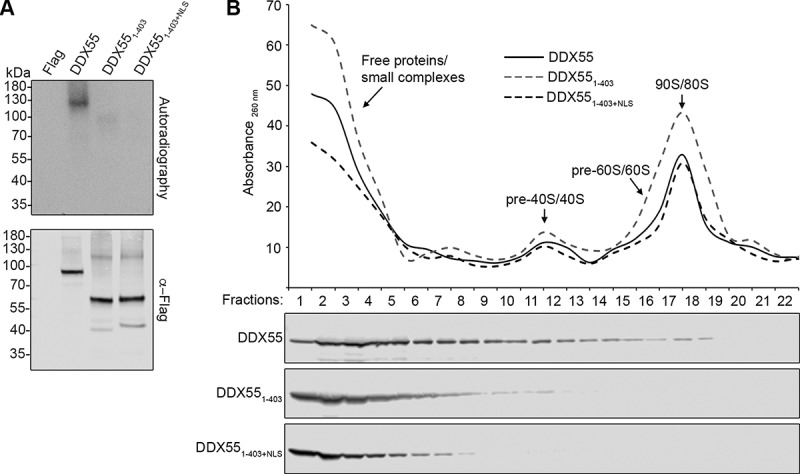
The C-terminal region of DDX55 is required for pre-ribosome association. (A) HEK293 cells expressing the Flag tag alone, DDX55-Flag, DDX551-403-Flag or DDX551-403+NLS-Flag were crosslinked using light at 254 nm. The tagged protein and crosslinked RNAs were retrieved on α-Flag beads under native conditions and subjected to a partial RNase digest. Complexes were then immobilized on NiNTA in denaturing conditions and co-purified RNA fragments were [32P] labelled. After elution, protein-RNA complexes were separated by denaturing PAGE, transferred to a nitrocellulose membrane and radiolabelled RNAs were detected by autoradiography. Protein samples were separated by SDS-PAGE and subjected to western blotting using an α-Flag antibody. (B) Whole cell extracts prepared from HEK293 cells expressing HEK293 cells expressing DDX55-Flag, DDX551-403-Flag or DDX551-403+NLS-Flag were separated by sucrose density gradient centrifugation. During fractionation, an absorbance profile at 260 nm was generated and the peaks corresponding to 80S mature/90S pre-ribosomes, and (pre-)40S and (pre-)60 particles are indicated (upper panel). Proteins in each fraction were separated by SDS-PAGE and analysed by western blotting using an α-Flag antibody (lower panel)
