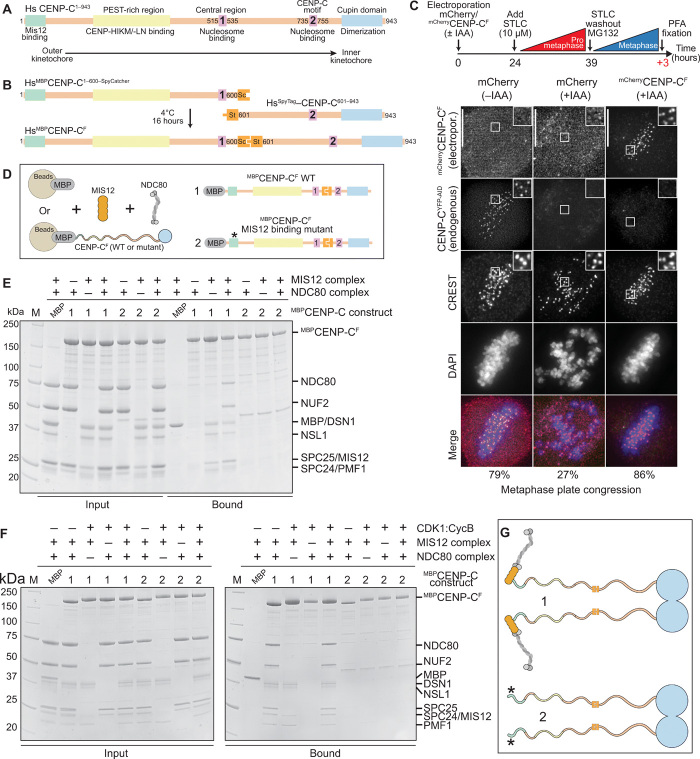
Fig. 1. Construction and validation of a CENP-C fusion protein.
(A) Organization of human CENP-C. The N-terminal MIS12 binding region is highlighted in green, and the CENP-HIKM/-LN binding region is highlighted in yellow. The central and C-terminal CENP-C motifs are highlighted in magenta. The C-terminal Cupin-like dimerization domain is highlighted in blue. The arrangement of the binding sites recapitulates the outer-to-inner kinetochore axis. (B) Strategy to purify full-length CENP-C using the SpyCatcher-SpyTag system. The two individual CENP-C fragments, CENP-C1–600–SpyCatcher and SpyTag–CENP-C601–943, were incubated together to form the full-length CENP-C ligation product. Sc, SpyCatcher; St, SpyTag. (C) Representative images showing fluorescence of electroporated mCherry or mCherryCENP-CF and endogenous CENP-CYFP-AID. Centromeres were visualized by CREST sera, and DNA was stained by 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm. Experimental regime: 24 hours after electroporation, cells were treated with 10 μM STLC for 15 hours. STLC (S-trityl-L-cysteine) was washed out three times, and 10 μM MG132 was added for 3 hours before paraformaldehyde (PFA) fixation. (D) Schematic of the performed amylose-resin pull-down assays. Maltose-binding protein (MBP) and two MBPCENP-CF variants, wild-type (WT) (1) and MIS12 binding mutant (2), were immobilized on amylose resin as bait. MIS12C and NDC80C were added as preys. (E and F) Result of the amylose-resin pull-down experiment. The MBPCENP-CF variant used as bait is indicated above each lane. MIS12C and NDC80C were added as indicated above each lane. MBPCENP-CF was additionally phosphorylated by CDK1:Cyclin B as indicated above each lane in (F). M, protein marker. (G) Graphical summary of the results shown in (E) and (F).