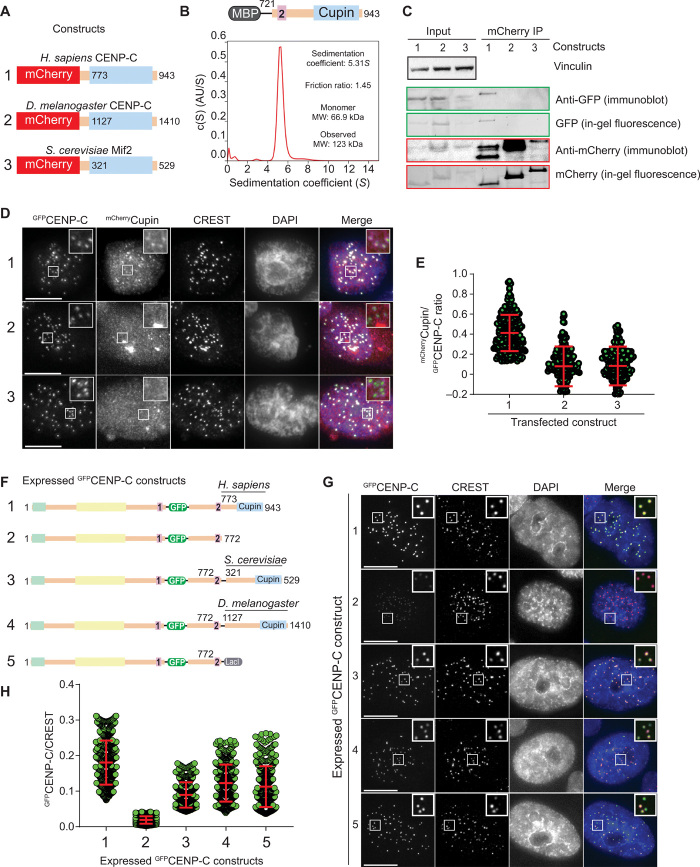
Fig. 2. CENP-C dimerization promotes kinetochore localization efficiency.
(A) Schematic showing the three mCherry-tagged Cupin constructs from Homo sapiens, S. cerevisiae, and D. melanogaster that were coexpressed with human GFPCENP-C in HeLa cells. (B) Sedimentation coefficient distribution obtained from the sedimentation velocity AUC experiment using MBPCENP-C721–943. AU, arbitrary unit. The observed molecular weight (MW) of 123 kDa indicates that the sample forms a dimer. (C) Western blot showing the results of coimmunoprecipitation (IP) experiments using RFP-Trap beads. (D) Representative images showing the fluorescence of GFPCENP-C and three different mCherryCupin fragments as shown in (A). Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm. (E) Quantification of the fluorescence intensities showing the mCherry/green fluorescent protein (GFP) ratio. Centromeres were detected in the CREST channel using a script for semiautomated quantification. Shown are means and SD from three independent experiments. (F) Schematic showing the different GFPCENP-C variants that were expressed in HeLa cells. (G) Representative images showing the fluorescence of the different GFPCENP-C variants as shown in (F). Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm. (H) Quantification of the GFP fluorescence intensities. Shown are means and SD from three independent experiments.