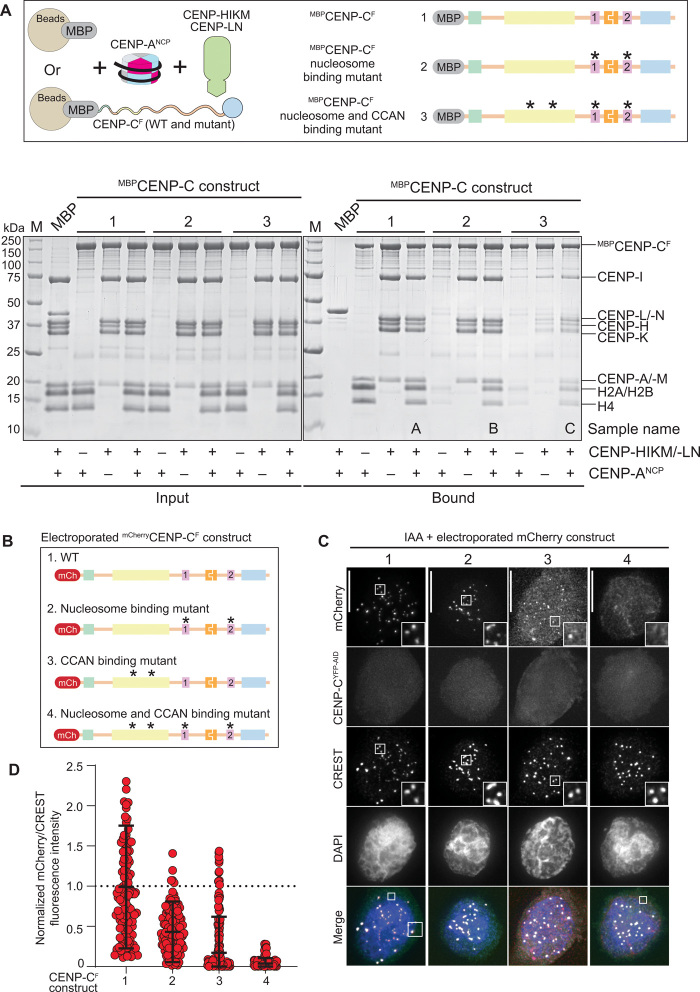
Fig. 4. Relative contributions of CENP-C and CCAN to CENP-A binding.
(A) Schematic of the performed amylose-resin pull-down assays. MBP and three MBPCENP-CF variants, WT (1), nucleosome binding mutant (2), and nucleosome and CCAN binding mutant (3), were immobilized on amylose resin as bait. CENP-ANCP and the CCAN members CENP-HIKM/-LN were added as prey. The two SDS-PAGEs show the results of the amylose-resin pull-down experiment. The MBPCENP-CF variant used as bait is indicated above each lane. CENP-ANCP and CENP-HIKM/-LN was added as indicated below each lane. The left gel shows the input fractions, and the right gel shows the bound fractions. CCAN binding mutant: L265A, F266A, L267A, W317A, E302A, F303A, I305A, and D306A (12, 14). Nucleosome binding mutant: R522A, W531A, R742A, and W751A. (B) Schematic showing the different electroporated mCherryCENP-CF variants. (C) Representative images showing the fluorescence of endogenous CENP-CYFP-AID and the electroporated mCherryCENP-CF variants as shown in (B). Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm. (D) Quantification of the mCherry fluorescence intensities normalized to the centromere marker CREST and to mCherryCENP-CF-WT. Shown are means and SD.