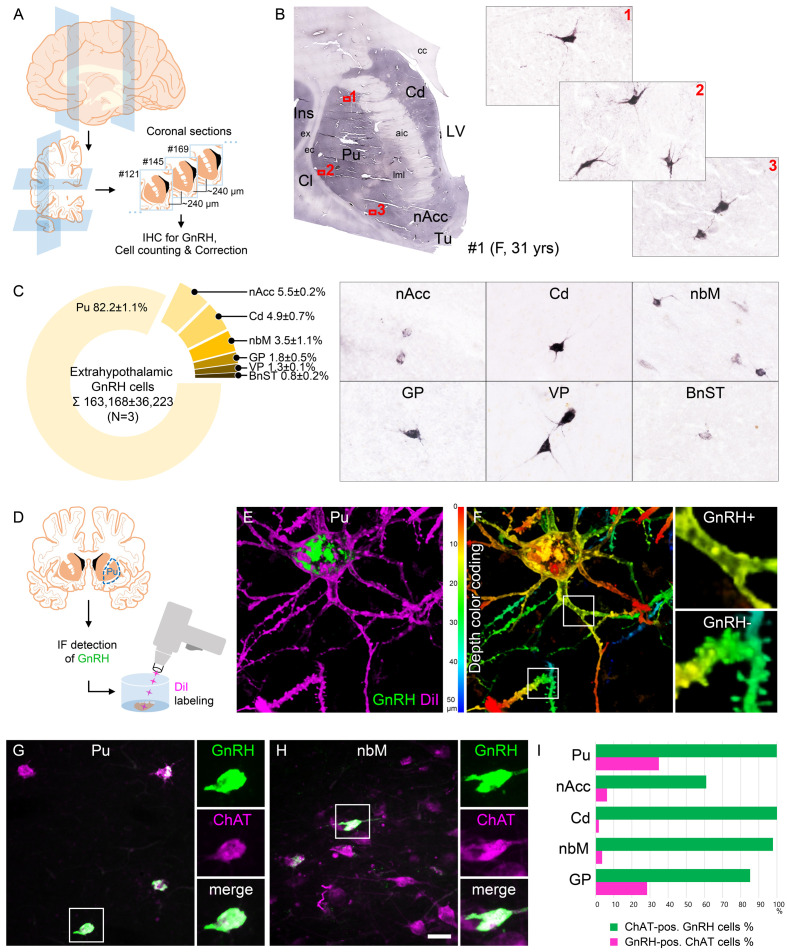
Figure 1. Anatomical approaches unveil the distribution, number, fine structure, and cholinergic phenotype of extrahypothalamic gonadotropin-releasing hormone (GnRH) neurons in the adult human brain.
(A) Extrahypothalamic GnRH-immunoreactive (GnRH-IR) neurons were mapped with immunohistochemistry (IHC) and quantified in the brain of three adult human individuals (#1–3). (B) Representative coronal section of a 31-year-old female subject (#1) illustrates the caudate nucleus (Cd), putamen (Pu), claustrum (Cl), insular cortex (Ins), anterior limb of the internal capsule (aic), external capsule (ec), extreme capsule (ex), corpus callosum (cc), lateral medullary lamina (lml), lateral ventricle (LV), nucleus accumbens (nAcc), and olfactory tubercle (Tu). High-power insets (1–3) reveal extrahypothalamic GnRH neurons many of which can be found in the Pu. (C) The majority (82.2%) of the 163,168 ± 36,223 extrahypothalamic GnRH neurons occurred in the Pu, followed by the nAcc, Cd, nucleus basalis magnocellularis (nbM), globus pallidus (GP), ventral pallidum (VP), and bed nucleus of the stria terminalis (BnST). (D) To visualize the fine structure of dendrites, the immunofluorescent (IF) detection of GnRH was combined with cell membrane labeling using DiI delivered with a Gene Gun. (E) 3-D reconstruction of the DiI-labeled (magenta) GnRH-IR (green) neurons revealed large multipolar cells, which exhibited only few dendritic spines. (F) Depth color coding, where colors represent distance from the section surface, allowed better distinction between DiI-labeled processes of the GnRH neuron (upper inset; GnRH+) from other DiI-labeled neuronal elements many of which belonged to medium spiny GABAergic projection neurons (lower inset; GnRH-). (G) Double-IF experiments addressed the presence of the cholinergic marker enzyme choline acetyltransferase (ChAT) in GnRH neurons. Nearly all GnRH neurons in the Pu contained ChAT signal. (H) The GnRH neuron population also overlapped with cholinergic projection neurons of the nbM. (I) With few exceptions, GnRH neurons were ChAT-immunoreactive (green columns), whereas they represented relatively small subsets of cholinergic cells (magenta columns) being the highest in the Pu (~35%). Scale bar (shown in H): 3.5 mm in low-power photomicrograph (B), 50 μm in (B, C, G and H) (insets in G and H: 25 μm), 12.5 μm in (E) and (F) (insets: 3.7 μm).