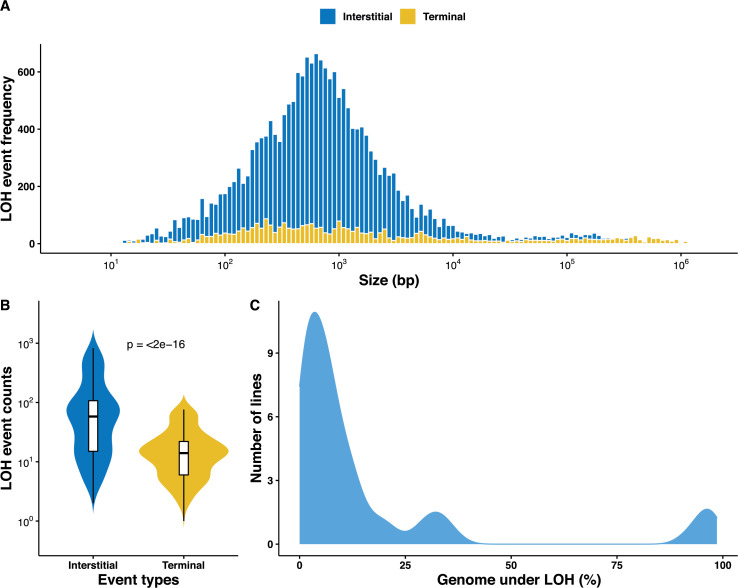
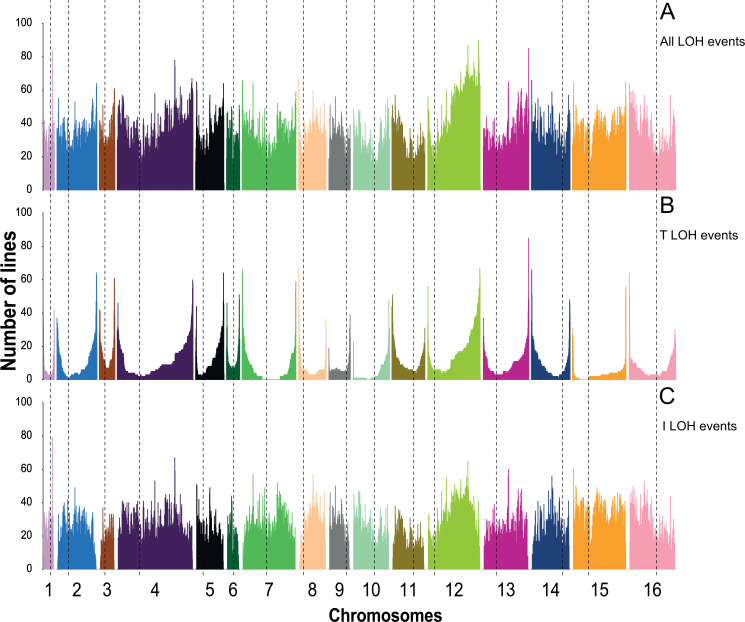
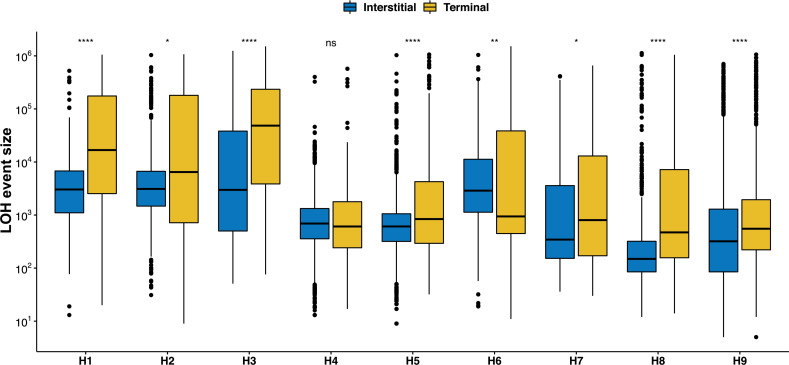
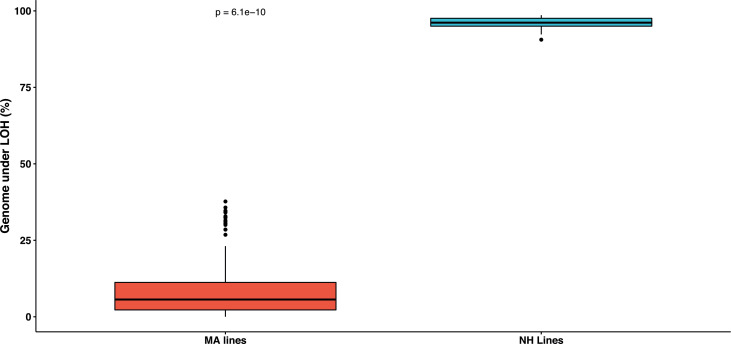
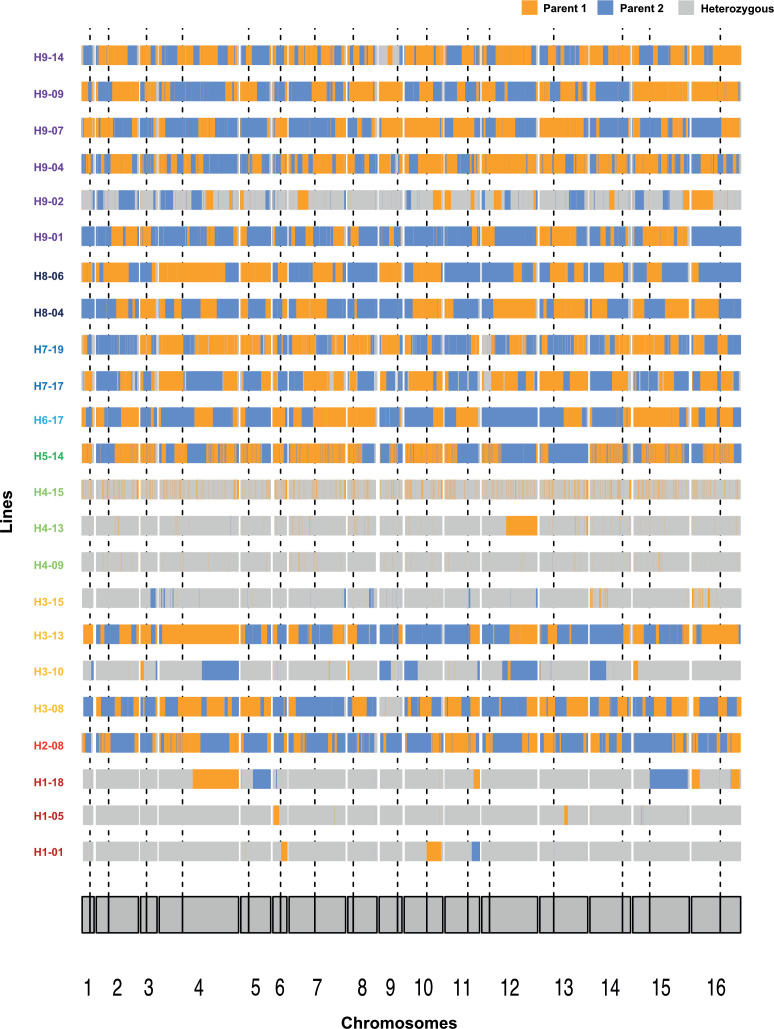
Figure 1. Overall distribution of loss of heterozygosity (LOH) in the 169 mutation accumulation (MA) lines.
(A) LOH event tract size distribution across all 169 MA lines, the average tract sizes of the interstitial LOH (I-LOH) events (7.4 kb) and terminal LOH (T-LOH) events (55.3 kb), respectively. The global average LOH event size was 14.1 kb. (B) Violin plot of the LOH event counts in the MA lines population, I-LOH events were found to be significantly greater than T-LOH events (Wilcoxon test, p<2×10−16). (C) Distribution of MA lines based on the proportion of genome under LOH (%), dashed line indicates average proportion of genome under LOH across the 169 MA lines, 15.9% (±1.86).