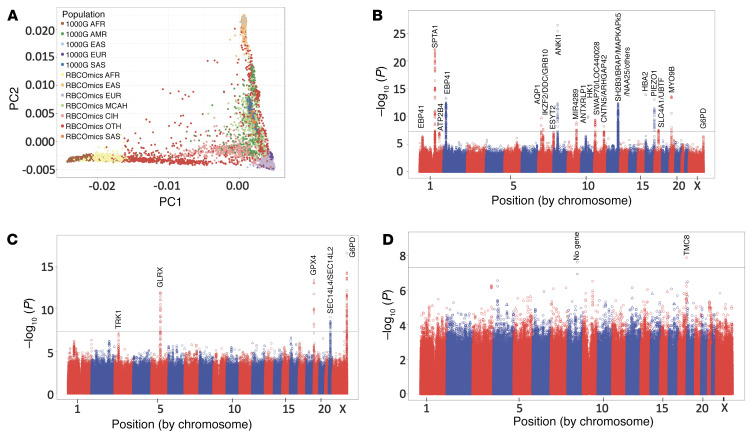
Figure 1. Ancestry of RBC-Omics population and Manhattan plots.
(A) Plot of the first 2 principal components (PCs) of the extended RBC-Omics population overlaid on the 1000 Genomes phase 3 samples. Individuals are labeled by genetic ancestry (AFR, African American; EAS, East Asian; SAS, South Asian; EUR, non-Hispanic White; AMR, admixed American; CIH, Caribbean Island Hispanics; MCAH, Mexican and Central American Hispanics; OTH, other/multiple ancestry) overlain by ancestry groups from 1000 Genomes v3. (B–D) Manhattan plots summarizing the mega-analysis results for osmotic hemolysis (n = 12,215, λ = 1.003; B), oxidative hemolysis (n = 10,007, λ = 1.048; C), and storage hemolysis (n = 12,177, λ = 1.002; D). Each data point corresponds to a –log10(P value) from a multivariant linear regression model’s P value for an SNP. The black horizontal line represents an accepted P-value level of genome-wide significance (P = 5 × 10–8). Circles represent noncoding variants, and triangles are coding variants.