Figure 3 |. BRCA1/Dicer/Ago promote sdRNA production; PalB2/Rad52 promote sdRNA-mediated repair.
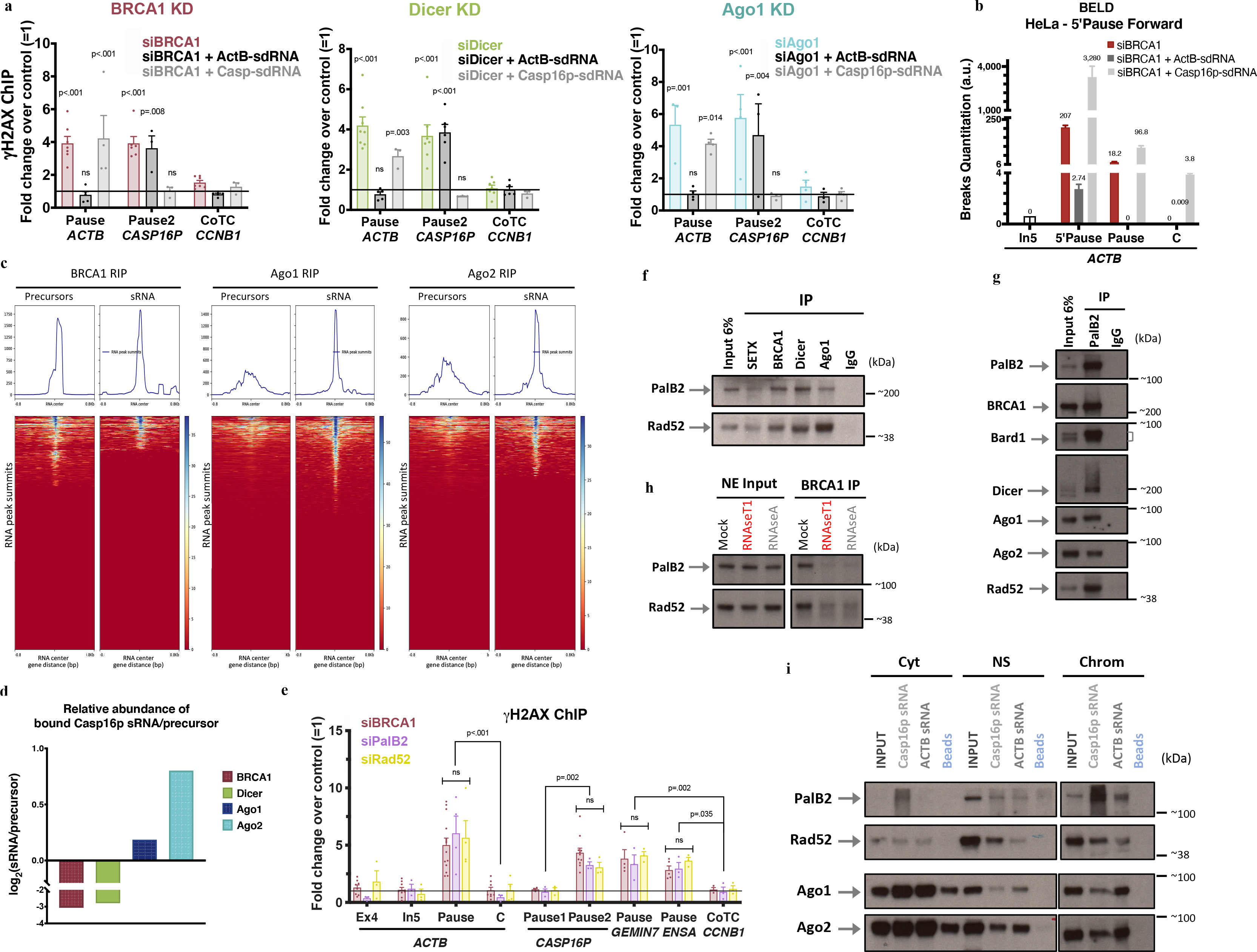
a, γ-H2AX ChIP analyses of ActB- and Casp16p-sdRNA complementation experiments in BRCA1-, Dicer- or Ago-depleted HeLa cells (n = 3–7; 3–7 and 3–4 biological replicates, respectively). b, Representative forward strand ACTB pause site BELD analysis in BRCA1-depleted HeLa cells, complemented with ActB- or Casp16p-sdRNA. Average fold change of qPCR replicates ± s.d. c, BRCA1, Ago1, and Ago2 genome-wide RNA RIP analyses of large (<300nt) and sRNAs, are shown as heat maps centered around the sdRNA peaks (+/− 0.8kb). d, HeLa WCE BRCA1, Dicer or Ago1/2 RIP analyses showing the binding ratio (Log2) between the Casp16p-sdRNA and its precursor. e, γ-H2AX ChIP DNA damage quantitation in PalB2- or Rad52-depleted HeLa cells. All ChIP analyses depict the average fold change ± s.e.m over the mock siRNA condition (n = 3–14 biological replicates). f-g, Immunoblots showing the proteins co-IPed from endogenous SETX, BRCA1, Dicer or Ago1 (f) and PalB2 (g) from HeLa NE, n=7. h, BRCA1 NE co-IP showing Rad52 and PalB2 immunoblots pre or post exposure to RNaseA or RNaseT1, n=6. i, RNA/protein binding assays, performed in HeLa subcellular fractions, using synthetic biotinylated Casp16p- and ActB-sdRNAs or streptavidin beads, were detected by immunoblotting (n=5). Cyt, cytoplasm; NS, Nuclear Soluble; Chrom, chromatin. ChIP data were all analyzed by Two-way ANOVA with post-hoc Tukey HSD and compared to the undamaged locus, or relevant control cells.
