Figure 4 |. Biological outcome of the interplay between sdRNA, PalB2 and Rad52 in pause site DNA damage repair.
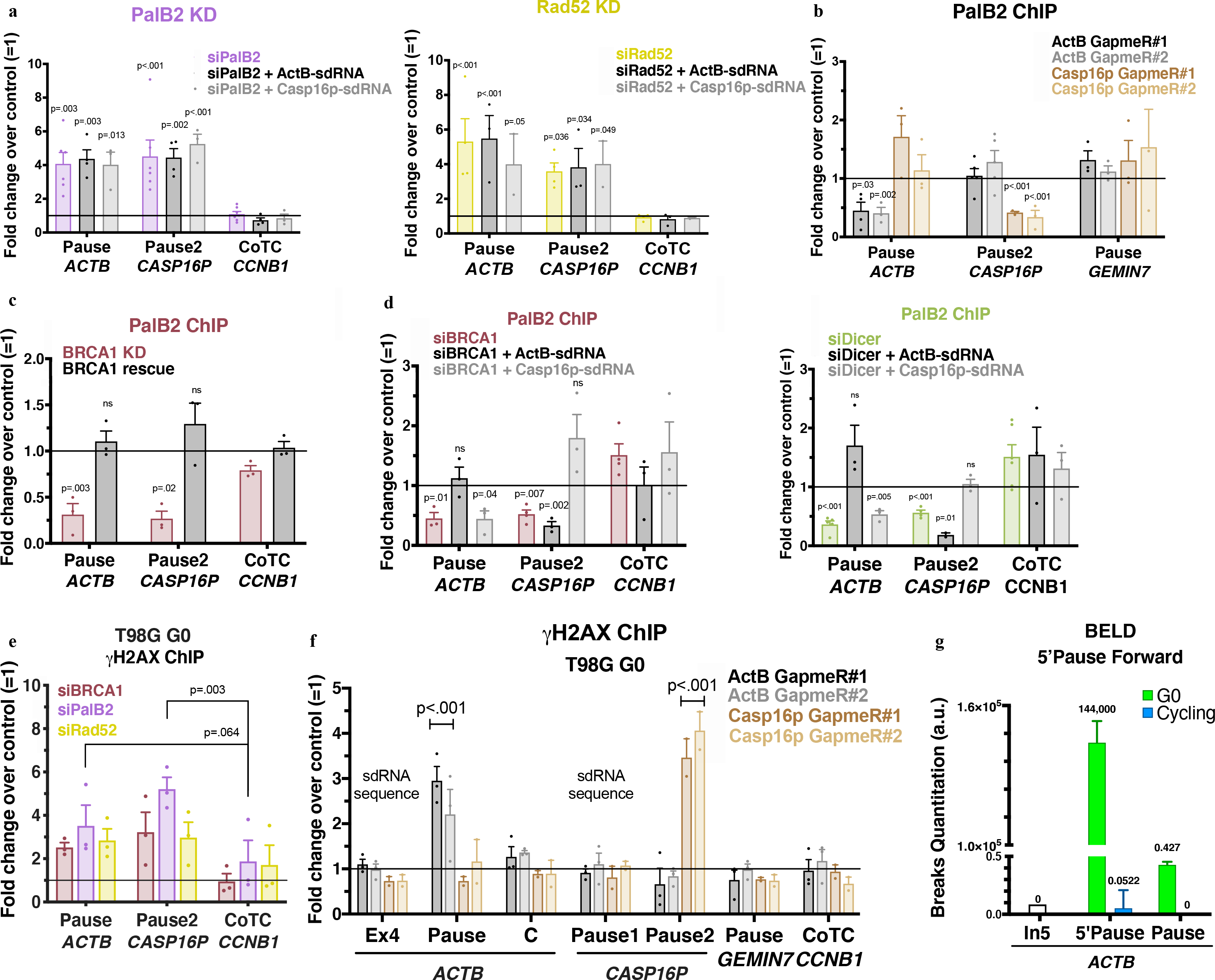
a, γ-H2AX ChIP analyses of ActB- and Casp16p-sdRNA complementation experiments in PalB2- or Rad52-depleted cells. b, PalB2 ChIP analyses with or without depletion of GapmeR-mediated ActB- or Casp16p-sdRNA. c, PalB2 ChIP analyses showing BRCA1-dependent PalB2 recruitment (n=3) d, PalB2 ChIP analyses of ActB- and Casp16p-sdRNA complementation experiments in BRCA1- or Dicer-depleted HeLa cells (n= 3–4 biological replicates). e, γ-H2AX ChIP quantitation analyzed in G0 BRCA1-, PalB2- or Rad52-depleted T98G cells (n= 3 biological replicates). f, γ-H2AX ChIP analyses in G0 T98G following ActB- or Casp16p-sdRNA GapmeR-mediated depletion (n = 3–4 biological replicates). g, ACTB forward strand BELD signal in quiescent (G0) and cycling T98G cells. Representative experiment showing the relative average abundance of the qPCR replicates ± s.d. ChIP analyses depict the average fold change ± s.e.m of the relevant depletion compared to the mock siRNA or negative GapmeR. Data were all analyzed by Two-way ANOVA with post-hoc Tukey HSD (panels c-e, multiple t-test) and compared to the undamaged locus, or relevant control cells.
