Extended Data Figure 1 |. BRCA1 interaction with RNAi factors.
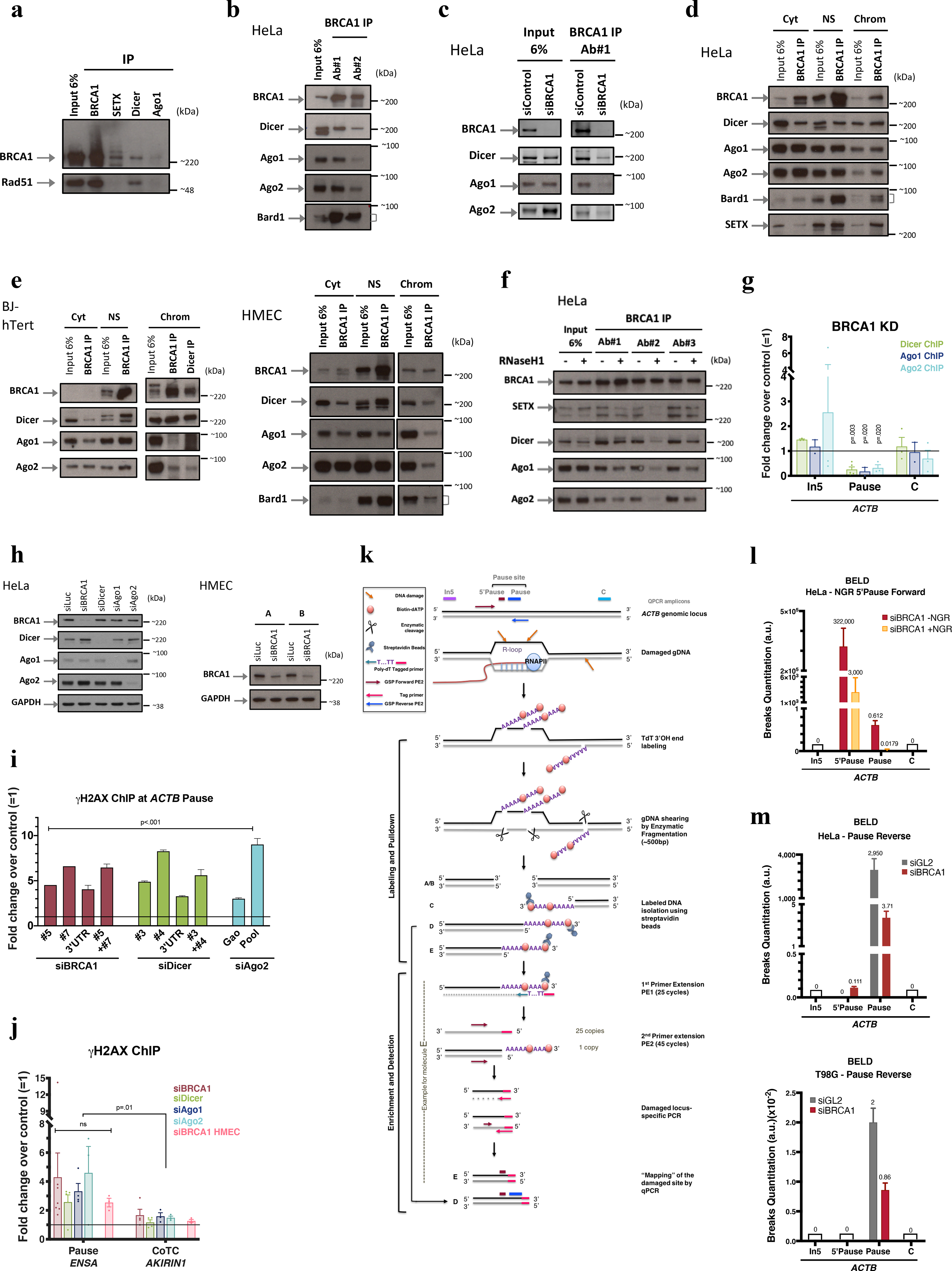
a, Co-IP of endogenous SETX, BRCA1, Dicer or Ago1 in HeLa NE showing that RAD51, an established BRCA1 binding partner involved in HR, failed to co-IP with SETX and co-IP’d very weakly, if at all, with Dicer and Ago1. n=3. b, Endogenous BRCA1 co-IP in HeLa NE, using two, BRCA1 Abs, each recognizing a different epitope (#1: N-terminus and #2: C-terminus). c, BRCA1 co-IP performed in siBRCA1-transfected HeLa NE; n=3 d-e, Co-IP of endogenous BRCA1 and RNAi factors present in subcellular HeLa fractions (d) and confirmed in HMEC and human fibroblast (BJ-hTert)(e) n=4. f, BRCA1 was co-IP’d, using three, monospecific Abs, in HeLa NE from cells overexpressing (+) or not (−) RNAseH1. Immunoblots showed with Ab#2 that a BRCA1/SETX/RNAi complex was R-loop sensitive (co-IP). IgG, negative control; Cyt, cytoplasm; NS, Nuclear Soluble; Chrom, chromatin. n=2 g, Dicer, Ago1, and Ago2 ChIP were performed in BRCA1-depleted HeLa,. Their ACTB pause site recruitment was BRCA1-dependent (n = 2–5 biological replicates). h, Representative immunoblots demonstrating siRNA-mediated silencing efficiency in HeLa cells and HMECs; n=10. i. Representative DNA damage analysis using individual siRNAs for each relevant target. j, DNA damage analyses at ENSA (R-loop-positive) and AKIRIN1 CoTC (R-loop-negative) transcription termination sites (n = 3–7 biological replicates). k, Broken End Labeling and Detection (BELD) strategy used to identify DNA breaks at the ACTB pause site. l-m, BELD quantitation of breaks at the ACTB pause site. l, BELD performed at the ACTB pause site in BRCA1- depleted cells with or without prior in vitro Nick and Gap Repair (+/− NGR) of breaks performed prior to TdT labeling. m, BELD performed on the ACTB pause site reverse strand. A representative example of each BELD analysis is shown, and the histograms depict the average values of qPCR replicates ± s.d. All ChIP data are represented as an average fold change ± s.e.m of the relevant KD condition compared to the mock. Data in g, i and j were respectively analyzed by multiple t-test, Two-way ANOVA with post-hoc Tukey HSD and unpaired t-test, and compared to results obtained with the undamaged locus or with relevant control cells.
