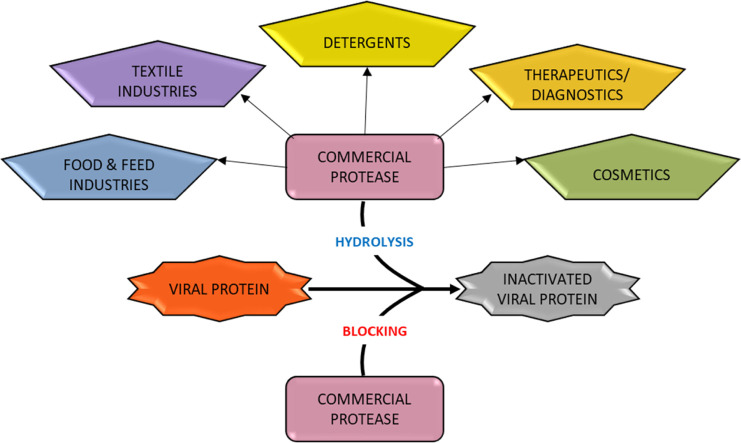
Abstract
Proteases have long been the target of many drugs, but their potential as therapeutic agents is a well-known, but under-explored area. Due to the heightened threat from new and emerging infectious agents, it is worthwhile to tap into the vast microbial protease resource to identify potential therapeutics. By docking proteases of the fungus Penicillium janthinellum NCIM 1366 with the proteins encoded by the SARS-CoV-2 virus, the enzymes that have the potential to bind with, and thereby degrade viral proteins were identified. In-silico docking analysis revealed that both fungal and commercially available proteases belonging to the A1A, M20A, S10, S8A and T1A families were able to bind the viral spike, envelope, ORF-7a and Nsp2 proteins (binding energy < −50 kJ/mol), thereby opening up the possibility of developing additional therapeutic applications for these enzymes.
Keywords: Penicillium, Fungal, Protease, Antiviral, Therapeutic, COVID
Graphical abstract
1. Introduction
In keeping with global efforts to reduce dependency on potentially hazardous chemicals and chemical processes, there has been a gradual shift towards establishing biological and biochemical processes to replace them. Various enzymes, especially proteases, are now used in diverse industries like textiles, detergents, leather, feed, pharmaceuticals, bioremediation etc. Proteases are one of the most widely used classes of industrial enzymes, accounting for up to 20% 0f the enzymes marketed worldwide (Singhal et al., 2012). Because of their versatility in performing both synthetic and degradative functions, proteases enjoy a ubiquitous distribution in nature. In particular, microbial proteases have excellent potential for commercial applications because of their robust nature and tolerance to harsh conditions.
Microbes account for nearly two-thirds of the commercial proteases produced worldwide (Beg and Gupta, 2003). Microbial proteases are usually extracellular, which simplifies their purification and other downstream processes (Nisha and Divakaran, 2014). In comparison, production of proteases from animals and plants are more labor-intensive. Additionally, owing to their broad-spectrum biochemical variety, higher yield, lower time consumption, lesser space requirement, ease of genetic manipulations and cost-effectiveness, microbes are the preferred source for commercial proteases (Ali et al., 2016).
Among the microbes, Bacillus sp. are extensively studied for protease production on a large scale; other proficient producers include Pseudomonas and Streptomyces sp. Fungal species like Aspergillus, Penicillium, Rhizopus, Mucor and Endothia, have been studied thoroughly for the production of acid, neutral and alkaline proteases. In spite of usually having lower reaction rates and lesser heat tolerance than their bacterial counterparts, one of the advantages of fungal proteases is that they can conveniently be produced in a solid-state fermentation process (da Silva, 2017).
Proteases, either as plasma fractions or as purified proteins, have clinically been used for the treatment of cardiovascular diseases, sepsis, digestive disorders, cystic fibrosis, inflammation, retinal disorders etc. (Craik et al., 2011). In addition to these, proteases are also used in the pharmaceutical and cosmetic industries for the removal of keratinized skin, depilation, vaccine preparation for dermatophytosis therapy, and for improved ungual drug delivery (Brandelli et al., 2010). Collagenolytic proteases have also been directly employed for wound healing, treatment of retained placenta, treatment of sciatica in herniated discs, and as a pretreatment for enhancing adenovirus-mediated cancer gene therapy (de Souza et al., 2015).
In case of viral infections, to date, nearly 20 different chemotherapeutic agents (that are mostly nucleoside analogs) have been approved for treatment via the inhibition of viral DNA synthesis/reverse transcription. These chemicals are used primarily for alleviating infections caused by herpes virus, the human immunodeficiency virus, respiratory syncytial virus and the influenza A virus (Burrell et al., 2017). However, most of these agents have limited clinical efficacy, adverse side effects, and suboptimal pharmacokinetics, which results in the use of chronic therapy that in turn leads to the emergence of drug-resistant viral strains that limit subsequent treatment options.
In the last 20 years, there has been seven significant viral threats – Nipah virus, SARS, MERS, Ebola, avian influenza, swine flu and now the SARS-CoV-2 mediated COVID-19. Estimates place about 60% of infectious diseases and 70% of emerging human infections as zoonotic in origin, with two-thirds originating in wildlife (Vorou et al., 2007). Due to human encroachment on the natural world, experts worldwide agree that this pandemic will not be the last; there exists the ideal conditions for diseases from wildlife to spill over into humans and spread quickly around the world.
To date, the global COVID-19 pandemic has caused 3.48 million deaths worldwide, including more than 300,000 deaths in India. The number of Indians who have/had infections stand at nearly 27 million. In addition to its devasting effects on healthcare systems worldwide, reports from the World Bank estimated that an outbreak of this scale could push about 49 million people into extreme poverty- almost half whom will be in Sub-Saharan Africa, with an additional 16 million in South Asia. In India, due to factors like where they live, where they work, high dependence on public services and limited savings and unavailability of insurance, it is estimated that 260 million people will be back in poverty due to the pandemic, from which approximately 40 million people will be in “extreme poverty” (Blake and Wadhwa, 2020).
As such, it is vital that adequate and affordable prevention/treatment options are in place to mitigate the adverse effects of such pandemics on livelihoods, healthcare and other public systems. Due to these concerns, the development of new antiviral agents is warranted, that can be used either as an alternative treatment strategy or as part of a combinational therapeutic approach (Patick and Potts, 1998).
Since recent technological advances have facilitated greater understanding of the essential viral enzymes, these proteins represent potential therapeutic targets. Because of the ease with each microbial proteases can be obtained, the recognition that proteases are an established class of safe and efficient drugs, and the fact that industrial scale processes already exist for the commercial production of many of them, it is worthwhile to explore their potential in degrading the viral enzymes, and thereby unearth additional therapeutic applications for these enzymes.
The overall objective of this study is to identify microbial proteases that have the potential to bind with, and degrade, viral proteins. Since microbial proteases are grouped into 83 families based on structure, functionality and substrate specificity, and as each family contains hundreds of entries, it will be a herculean task to analyze representatives of each subgroup. Therefore, to reduce the size of the dataset, the proteins of the fungus Penicillium janthinellum NCIM 1366 (henceforth referred to as PJ-1366) have been used as a model group, and the proteins encoded by SARS-CoV-2 has been used as a representative of enveloped viral proteins.
2. Materials and methods
2.1. Prediction of secreted proteases from PJ-1366, annotation and ontology prediction
From the whole genome sequence of PJ-1366 obtained by paired end sequencing, genes were predicted using AUGUSTUS (Stanke et al., 2004). From the list of predicted genes, proteins with signal sequences were identified using SignalP- 5.0 (Almagro Armenteros et al., 2019).
A database of proteases reported from Penicillium sp. was created using information available on the MEROPS database (Rawlings et al., 2018). A sequence similarity search between the extracellular proteins of PJ-1366 and the custom peptidase database was performed using blastp. The resulting matches were annotated using the UniProt KB (Magrane and Consortium, 2011). Any sequence not annotated as a protease was removed from the list.
The remaining entries were processed using the BlastKOALA tool (Kanehisa et al., 2016) to identify sequences reported as peptidases/inhibitors in the KEGG BRITE database.
2.2. Homology modelling of predicted inhibitors
The SWISS-MODEL server (Biasini et al., 2014) was used for homology modelling of the predicted protease inhibitors. Wherever possible, models with the highest GMQE (Global Model Quality Estimation) and QMEAN Z-scores above -4.0 were selected for further analysis.
2.3. Molecular docking and analysis
The structures of the proteins of the SARS-CoV-2 virus were obtained from the UniProt databank. Using PJ-1366 proteases as the receptor and SARS-CoV-2 proteins as the ligand, molecular docking based on shape complementarity principles was performed using PatchDock (Schneidman-Duhovny et al., 2005) with default parameters and clustering RMSD of 4.0. The docking solutions were refined and scored according to energy function using FireDock (Mashiach et al., 2008).
2.4. Analysis of binding affinities of related microbial proteases
The structures of other microbial proteases belonging to the same families as PJ-1366 proteins with good binding potential (−50 to −80 kcal/mol) to viral proteins were selected from the MEROPS database, and their binding affinities to SARS-CoV-2 proteins were studied using PatchDock and FireDock.
3. Results and discussions
3.1. Prediction of secreted proteases from PJ-1366, annotation and ontology analysis
From the 37.6 Mbp genome of PJ-1366, 11,828 genes were predicted by AUGUSTUS. Out of these, 1007 sequences had putative eukaryotic signal sequences as per SignalP analysis.
From the MEROPS peptidase database, a custom list of 2146 proteases from 49 Penicillium species was created. Blastp alignment gave 175 PJ-1366 proteins with significant similarity. Based on UniProt annotation, 109 sequences were selected which were described as proteases/hypothetical proteins. BlastKOALA analysis of these sequences annotated 17 sequences as Peptidases and Inhibitors, based on BRITE hierarchical analysis (Table 1 ).
Table 1.
Orthology of sequences annotated as peptidases/inhibitors in the KEGG database.
| Orthology | Definition | No. of sequences |
|---|---|---|
| K01279 | Tripeptidyl-peptidase I [EC:3.4.14.9] | 5 |
| K01288 | Carboxypeptidase D [EC:3.4.16.6] | 4 |
| K01293 | Gly-Xaa carboxypeptidase [EC:3.4.17.4] | 1 |
| K01336 | Cerevisin [EC:3.4.21.48] | 1 |
| K01341 | Kexin [EC:3.4. 21.61] | 1 |
| K02739 | 20S proteasome subunit beta 2 [EC:3.4.25.1] | 1 |
| K05994 | Bacterial leucyl aminopeptidase [EC:3.4.11.10] | 1 |
| K08783 | Extracellular matrix protein 14 [EC:3.4.17.-] | 1 |
| K13289 | Cathepsin A (carboxypeptidase C) [EC:3.4.16.5] | 1 |
| K19305 | Deuterolysin [EC:3.4.24.39] | 1 |
From the orthology analysis, it was seen that some of the enzymes- such as tripeptidyl-peptidase 1, cerevisin and cathepsin, are lysosomal components where their natural function is protein degradation (He et al., 2015; Sohar et al., 2013), while others, like carboxypeptidase D, kexin and leucyl aminopeptidases, are involved in the pre-processing of proteins, especially hormones (Cawley et al., 2014; Fuller, 2013). Interestingly, one of the proteins matched to deuterolysin, which has long been used in the food industry, and is known usually to be thermostable (Maeda et al., 2016).
In the MEROPS database, proteases are grouped into 84 families according to their evolutionary relationship, under seven catalytic types: serine, aspartic, cysteine, threonine, glutamic acid and metallo-proteases, and asparagine peptide lyases (which catalyze via an elimination reaction rather than by hydrolysis). Using this information from MEROPS, the proteases/inhibitors of PJ-1366 were further sorted into different protease families based on structure-based classification. It was seen that serine proteases were the most prevalent (10 sequences), followed by metalloproteases (5 sequences) and one each of aspartic and threonine proteases.
Proteolytic enzymes are generally classified either based on the site of their action (as exopeptidases and endopeptidases), or by the optimal pH in which they are active (as acid/neutral/alkaline proteases). Based on UniProt descriptions (Table 2 ), it was seen that carboxypeptidases were the most prevalent. Since these enzymes generally are zinc-containing exopeptidases that remove single amino acids or dipeptides from the carboxyl end of oligopeptides, their applicability in destroying viral proteins may be limited. Consequently, proteases like the tripeptidyl peptidases, Penicillolysin (which is involved in degradation of proteins for nutrient uptake (Ichishima, 2004)), Aorsin and proteasome subunits are of more interest since, being endopeptidases, they can potentially disrupt viral protein structures by hydrolyzing peptide bonds in the interior of polypeptide chains.
Table 2.
UniProt description and MEROPS family classification of PJ 1366 proteases annotated as inhibitors.
| S. no. | Sequence ID | UniProt description | Length | Per. ident. | Family |
|---|---|---|---|---|---|
| 1 | ctg7180000009921.g250 | Penicillolysin [P. brasilianum] | 467 | 86.84 | M12B |
| 2 | ctg7180000009929.g55 | putative carboxypeptidase [P. rolfsii] | 585 | 91.70 | M20A |
| 3 | ctg7180000009963.g312 | Carboxypeptidase Y [P. rolfsii] | 581 | 95.11 | M20A |
| 4 | ctg7180000014270.g175 | serine carboxypeptidase [P. brasilianum] | 523 | 84.45 | S10 |
| 5 | ctg7180000014384.g300 | Proteasome subunit beta type-2 [P. subrubescens] | 356 | 98.56 | T1A |
| 6 | ctg7180000014413.g297 | Carboxypeptidase cpdS [P. brasilianum] | 524 | 90.50 | S10 |
| 7 | ctg7180000014422.g216 | Tripeptidyl-peptidase sed2 [P. subrubescens] | 607 | 90.83 | S53 |
| 8 | ctg7180000014558.g236 | Pheromone-processing carboxypeptidase kex1 [P. rolfsii] | 619 | 87.42 | S10 |
| 9 | ctg7180000014559.g244 | Tripeptidyl-peptidase sed4 [P. subrubescens] | 599 | 88.38 | S53 |
| 10 | ctg7180000015102.g232 | Sorting nexin-4 [P. rolfsii] | 1365 | 90.21 | S8B |
| 11 | ctg7180000015122.g39 | Leucine aminopeptidase 1 [P. brasilianum] | 390 | 93.25 | M28E |
| 12 | ctg7180000015128.g184 | putative metallocarboxypeptidase ecm14 [P. rolfsii] | 542 | 84.70 | M14A |
| 13 | ctg7180000015233.g14 | Aorsin [P. brasilianum] | 653 | 79.04 | S53 |
| 14 | ctg7180000015271.g77 | Serine-type carboxypeptidase F [P. subrubescens] | 540 | 86.49 | S10 |
| 15 | ctg7180000015281.g270 | Putative Protease S8 tripeptidyl peptidase I [P. brasilianum] | 676 | 89.81 | A1A |
| 16 | ctg7180000015289.g86 | Tripeptidyl-peptidase sed1 [P. subrubescens] | 656 | 89.94 | S53 |
| 17 | ctg7180000015373.g99 | Alkaline protease 2 [P. subrubescens] | 510 | 96.02 | S8A |
3.2. Homology modelling of predicted inhibitors
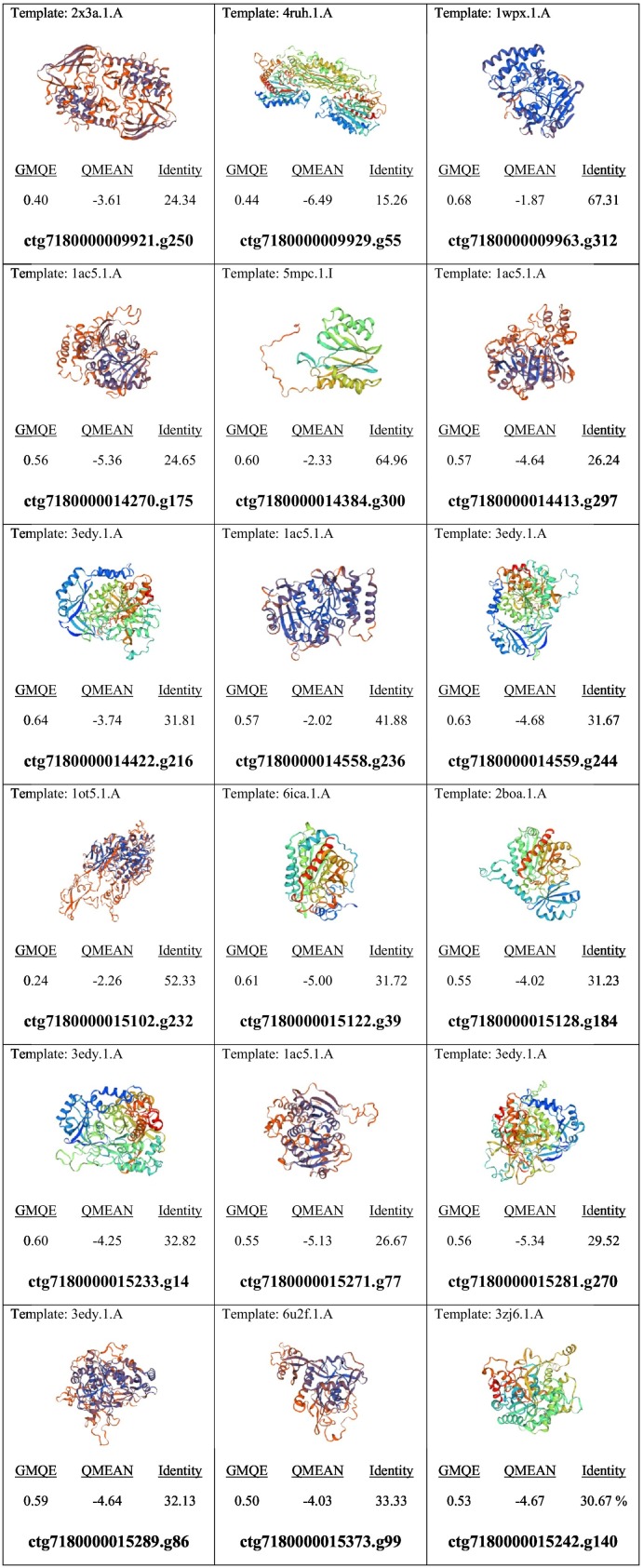
3D models of the selected PJ-1366 proteases were obtained by homology modelling from the SWISS-MODEL server. While GMQE values of all models were >0, 11 models had QMEAN < -4.00, indicating lower quality. Also, it was observed that multiple models were modelled on the same template, the most common of which were 1ac5- which is the kex1 delta-p subunit, and 3edy- which is a tripeptidyl peptidase 1 from Homo sapiens (Fig. 1 ). In yeast, the KEX1 protease is involved in apoptosis caused by defective N-glycosylation (Hauptmann and Lehle, 2008), while TPP1 in humans is found in lysosomes to digest and recycle different types of molecules (Stumpf et al., 2017).
Fig. 1.
Summary of homology models of selected PJ-1366 proteases.
From Fig. 1, it was seen that in most cases, the sequence identity was less than 50%. Better matches were obtained only for 3 sequences- ctg7180000009963.g312 (carboxypeptidase), ctg7180000014384.g300 (proteasome sub-unit) and ctg7180000015102.g232 (nexin). Nexin is a component of fibroblasts that links thrombin and plasminogen activator and mediates their binding to cells (Baker et al., 1980).
3.3. Molecular docking and analysis
Positive-strand RNA viruses like the SARS-CoV-2 virus are a group of related viruses that have positive-sense, single-stranded RNA genomes. The RNA genome acts as an mRNA that is translated into viral proteins using the host cell's ribosomes. Coronaviruses have the largest known RNA genomes, between 27 and 32 kilobases in length. Such viruses account for a significant fraction of known viruses. In humans and birds, these viruses are known to cause respiratory tract infections. Some of the coronaviruses cause mild illnesses in humans including some cases of the common cold, while more lethal varieties can cause SARS, MERS, and COVID-19.
In the case of SARS-CoV-2, each virion is 50–200 nm wide and has four structural proteins- the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins (Supplementary File 1). The N protein holds the RNA genome which is a single-stranded 29.9 kb long mRNA encoding 13 ORFs; the S protein allows attachment and fusion with the host cell membrane, the M proteins are responsible for virion morphogenesis, while the envelope small membrane proteins participate in ESCRT-independent budding for the formation of new virus particles (Neuman et al., 2011; ViralZone, 2020).
The SARS-CoV-2 genome encodes a ~7096 residue long polyprotein which consists of the structural and non-structural proteins (Tao et al., 2020; Wu et al., 2020). Expression of the viral proteins is either through a primary translation of the polyprotein that initiates infection, or after some replication, through sub-genomic mRNA expression which produces all structural proteins (Kim et al., 2020). A summary of the interactome of SARS-CoV-2 proteins with human proteins is provided in Supplementary File 2.
Eight viral proteins whose structures were resolved were used for docking- the Spike Glycoprotein (involved in viral attachment and entry), the ORF-7a Protein (which disrupts Tetherin antiviral activity), the Envelope protein (participates in viral budding), the Nucleoprotein (involved in viral genome packaging), the non-structural proteins Nsp1, Nsp2 and Nsp14 (which interfere with host cellular processes), and the ORF-6 protein (which disrupts interferon signaling by preventing nuclear import of proteins). Docking using PatchDock gave results sorted on the basis of the shape complementarity score, the interface area of the docked molecule, and atomic contact energy. The top 10 results were analyzed using FireDock, and the most favorable global binding energy of the complex was noted. The energies were color coded from red (most favorable) to green (least favorable) in order to better visualize the comparison between them (Fig. 2 ).
Fig. 2.
Binding energies of PJ-1366 proteases with SARS-CoV-2 proteins.
The energies are ranked from red (most favorable) to green (least favorable). Gene IDs of proteins with highly favorable binding energies (<-50 kcal/mol) are highlighted in yellow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
It was observed that while most of the non-structural viral proteins did not have favorable binding energies with PJ-1366 proteases, structural proteins like the spike glycoprotein and envelope protein, which are crucial for viral entry to host cells, were capable of being bound by the fungal proteases. Of the 17 fungal proteases that were analyzed, 7 structures showed favorable binding potential. These were ctg7180000009929.g55 (carboxypeptidase), ctg7180000014270.g175 (serine carboxypeptidase), ctg7180000014384.g300 (Proteasome subunit beta type-2), ctg7180000014413.g297 (Carboxypeptidase cpdS), ctg7180000015271.g77 (Serine-type carboxypeptidase), ctg7180000015281.g270 (Putative S8 tripeptidyl peptidase I) and ctg7180000015373.g99 (Alkaline protease 2).
Like bacteria, fungi are also susceptible to attack by viruses called mycoviruses, the majority of which are dsRNA viruses, though approximately 30% are +ssRNA viruses belonging to families like Barnaviridae, Narnaviridae, Pseudoviridae, Metaviridae etc. (May and Nowak, 1995). However, unlike in bacterial and mammalian systems, the mechanisms by which the fungal hosts tolerate or overcome these infections is not precisely known. Mycophages often have barely detectable effects on the host's fitness- a neutral co-existence that might be the result of co-evolution. Also, in certain yeasts and Ustilago species, retaining the virus actually proves to be beneficial as it increases the fungal pathogenicity (Pearson et al., 2009).
Even though their roles in overcoming mycoviral attacks is not known, several bioactive compounds, mostly polysaccharides, terpenoids and phenolics that are beneficial for human health have been derived from fungi, especially mushrooms (Seo and Choi, 2021). Reports on fungal proteins exhibiting antiviral effects are rare, though not unknown. At least three fungal proteins are known to inhibit the reverse transcriptase of the human immunodeficiency virus- a ubiquitin-like protein (Wang and Ng, 2000), a laccase (Wang and Ng, 2004) and a peptide inhibitor (Wang et al., 2007). Another protein- Nebrodeolysin- has been reported to inhibit HIV-induced syncytia formation in cells (Lv et al., 2009). Two other proteins- a laccase from Pleurotus ostreatus (El-Fakharany et al., 2010) and tyrosinases from Agaricus bisporus (Lopez-Tejedor et al., 2020) have also shown inhibitory effects against the Hepatitis C virus. However, there are no prior studies and reports on the antiviral efficacies of fungal proteases. Therefore, in addition to nutrient assimilation, whether any of the secreted proteases of PJ-1366 have roles in fighting viral infections is purely speculatory at this point.
3.4. Analysis of binding affinities of related microbial proteases
Proteases are architectural diverse- ranging from small enzymes (~20 kDa) to sophisticated multi-domain structures like proteasomes and meprin metalloproteinases (0.7–6 MDa). This multiplicity of enzymes results in an outstanding diversity in protease functions. Diversity is also observed in the case of specificity towards the targets, with some proteases exhibiting exquisite substrate/bond preferences; however, most proteases are relatively non-specific and can target multiple substrates (López-Otín and Bond, 2008).
Using information from the MEROPS family classification of peptidases, it was observed that the PJ-1366 proteases with the most favorable binding energies to viral proteins belonged to diverse families of endopeptidases- one aspartic protease (A1A), one metalloprotease (M20A), four serine proteases belonging to families S8A and S10, and a threonine protease (T1A) (Table 3 ).
Table 3.
Classification of fungal proteases with high binding affinity to viral proteins.
| Sequence ID | Family | Enzyme type | SWISS-MODEL template |
|---|---|---|---|
| ctg7180000015281.g270 | A1A | Pepsin A (Homo sapiens) | 2QZW Candida albicans |
| ctg7180000009929.g55 | M20A | Glutamate Carboxypeptidase (Pseudomonas sp.), | 1CG2 Pseudomonas sp. RS-16 |
| ctg7180000014270.g175 | S10 | Carboxypeptidase Y (Saccharomyces cerevisiae) | 1YSC Saccharomyces cerevisiae |
| ctg7180000014413.g297 | |||
| ctg7180000015271.g77 | |||
| ctg7180000015373.g99 | S8A | Subtilisin Carlsberg (Bacillus licheniformis) | 1SCD Bacillus licheniformis |
| ctg7180000014384.g300 | T1A | Archaean proteasome, beta component (Thermoplasma acidophilum) | 3H4P M. jannaschii |
Based on structure, the aspartic proteinases (APs) are classified into five superfamilies- AA, AC, AD, AE, and AF. The A1 family of eukaryotic APs is part of the AA clan, and contains many well-characterized enzymes with industrial and therapeutic uses (Rawlings et al., 2018). Not only do these peptidases hydrolyze proteins for nutrition and recycling, but they also perform many essential post-translational processing events for the activation/inactivation of enzymes and peptide hormones. Pepsin-like enzymes are aspartic proteases, which belong to the A1 family of peptidases. Pepsin hydrolyses proteins into water-soluble fragments called peptones. Partial digestion by pepsin has been commercially used for processing proteins in food industries. Medically. it has also been employed as a laxative (Summers, 2017).
Among the various peptidases in family M20 are carboxypeptidases such as the glutamate carboxypeptidase from Pseudomonas (M20.001), the thermostable carboxypeptidase Ss1 of broad specificity from archaea such as Sulfolobus sp. (M20.008) and the yeast Gly-X carboxypeptidase (M20.002). Bacterial glutamate carboxypeptidases- that have high affinity for folic acid- have been developed for anti-cancer regimes in two settings- to eliminate methotrexate from circulation rapidly, and to remove the glutamate residue from pro-drugs to release a cytotoxic agents at tumor sites.
The peptidase family S10 is active only at acidic pH, unlike most other serine peptidase families. Carboxypeptidase Y (CPY) is a glycoprotein exopeptidase with a broad amino acid specificity that can retain its activity under the denaturing conditions used for polypeptide sequencing (Nielsen et al., 1990). CPY has also been used as a sensing element in biosensors for the direct detection of ochratoxin A in olive oil (Dridi et al., 2015).
Most members of the family S8A are neutral/alkaline endopeptidases. Many peptidases in the family are thermostable. Because of this, these proteases, especially engineered subtilisins, have extensive applications in various industrial sectors such as detergent and leather industries, cosmetics, food processing, skin care ointments, metal scavenging and waste treatment (Sharma et al., 2019).
The ubiquitin–proteasome system participates in the regulation of most fundamental cellular processes via intracellular protein degradation. However, its proteolytic core, the 20S proteasome, has found to be attached also to the cell plasma membrane and certain observations suggest that they may be released into the extracellular medium (Sixt and Dahlmann, 2008). The eukaryotic proteasome has three different activities (trypsin-like, chymotrypsin-like and cleavage after glutamate). Each activity resides in a different β subunit. The archaean and bacterial proteasomes have only chymotrypsin-like activity, which are included in T1A. While the proteasome is an established anticancer drug target (Osmulski et al., 2017), utilizing the proteasome for degradation of heterologous proteins is an yet unexplored area.
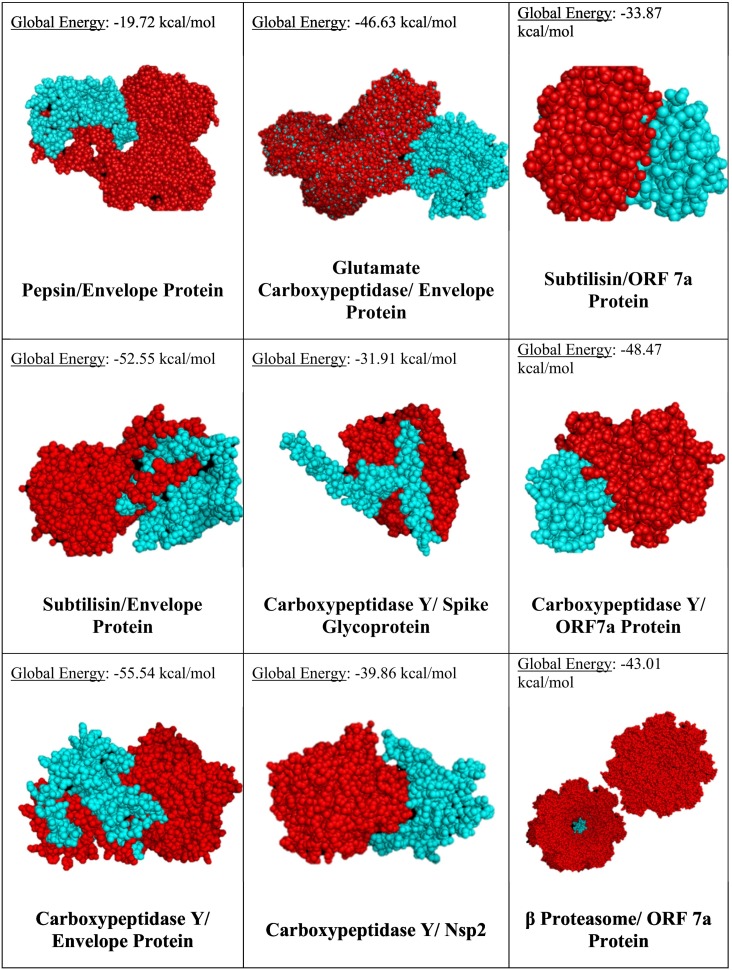
The structures of industrially-produced proteases from the five protease families with high binding potential were obtained from the MEROPS database, and were used for docking with viral proteins. After refinement of docking results using FireDock, it was seen that while all the proteases were able to bind the viral proteins in their catalytic pockets, the binding energies were noticeably higher (Fig. 3 ). This shows that in spite of being evolutionarily related, changes in amino acid sequences can still cause significant differences in the tertiary structure, which in turn affects the binding potential. Another factor to be reckoned with, is the imperfect modelling of the fungal proteases. Yet another possibility is that, due to evolution and adaptation, and their ability to colonize different environments and life forms, fungal proteins simply might have better chances of binding to viral proteins than the prokaryotic ones. This might be the reason for viral attacks not having highly deleterious effects on fungi.
Fig. 3.
Docking of commercial proteases (red) with viral proteins (cyan). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Due to the unprecedented and unchecked spread of the COVID-19 outbreak globally, current treatments have mostly focused on alleviating symptoms and providing respiratory support. The development of different vaccines against SARS-CoV-2 are either completed or nearing completion, and at least 13 different vaccines (across 4 platforms) are now being administered globally (Prompetchara et al., 2020). Still, the threat persists, mainly due to the rapidly evolving nature of the virus, and also due to the possible emergence of other zoonotic diseases. The situation is more dire in a country like India, with extreme geo-climatic and socio-economic diversity, and the nation faces a constant threat of emerging and re-emerging viral infections of public health importance. Therefore, repurposing and re-evaluating existing drugs and commercially available inhibitors against druggable targets of the virus could effectively accelerate the drug discovery process. In this regard, targeting essential proteins (viral and/or host) involved in viral entry and proliferation can be considered as a practical approach to alleviate the epidemic.
An important aspect to consider would be the specificity of the proteases against the targeted protein, since non-specific inhibition can adversely affect the regular physiological functioning of the host, either by activation of endogenous proteases or through degradation of protease inhibitors. Also, the proteases themselves can be immunogenic, and can induce inflammatory responses. For example, certain protease-activated receptors (PARs) have been known to alter the permeability of epithelial barriers which leads to inflammation (Enjoji et al., 2014). Some allergens also elicit an immune response through the protease-mediated cleavage of PARs, which induces proinflammatory cytokines and chemokines (Maeda et al., 2013).
Therefore, any assessment of a protease-based strategy should take into consideration the availability, effectiveness, safety and cost of alternative measures, including checking the spread of infection, immunization or treatment with existing drugs. In addition to in-vivo effects, the possibility of using these proteases as external antiviral agents, either to hydrolyze or competitively bind viral proteins, needs to be explored. New technologies for rationally engineering proteases, as well as improved delivery options, will significantly expand the efficacy of these enzymes.
4. Conclusions
Since proteases are already being used worldwide for a multitude of commercial applications, a combination of modelling studies was performed to identify proteases that could bind, and potentially degrade, SARS-CoV-2 proteins. Binding energy evaluation identified 7 proteases belonging to 5 different families that are suitable candidates for further evaluation. Based on our current understanding on the roles and physiological effects of proteases, it is proposed that a two-pronged clinical approach, aimed at either destroying or inhibiting viral proteins, could be applied for a more robust response against SARS-CoV-2, with due consideration given to the dosage and site of protease activity.
CRediT authorship contribution statement
Meera Christopher: Formal analysis, Investigation, Visualization, Writing of Original Draft. Prajeesh Kooloth-Valappil: Formal analysis, Validation, Review & Editing.
Athiraraj Sreeja-Raju: Formal analysis, Investigation.
Rajeev K. Sukumaran: Conceptualization, Methodology, Review & Editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
MC, PKV and ASR are thankful to the Council of Scientific and Industrial Research (CSIR), Govt. of India for the Junior/Senior Research Fellowships for Ph.D. studies. This study was partially funded by the Department of Biotechnology, Govt. of India (BT/PR20695/BBE/117/211/2016) and CSIR, Govt. of India (MLP 0035: 33/2018/MD-FTT&FTC-ANB).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biteb.2021.100756.
Appendix A. Supplementary data
Supplementary material
References
- Ali N., Ullah N., Qasim M., Rahman H., Khan S.N., Sadiq A., Adnan M. Molecular characterization and growth optimization of halo-tolerant protease producing Bacillus Subtilis Strain BLK-1.5 isolated from salt mines of Karak, Pakistan. Extremophiles. 2016;20:395–402. doi: 10.1007/s00792-016-0830-1. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019 doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Baker J.B., Low D.A., Simmer R.L., Cunningham D.D. Protease-nexin: a cellular component that links thrombin and plasminogen activator and mediates their binding to cells. Cell. 1980;21:37–45. doi: 10.1016/0092-8674(80)90112-9. [DOI] [PubMed] [Google Scholar]
- Beg Q.K., Gupta R. Purification and characterization of an oxidation-stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzym. Microb. Technol. 2003;32:294–304. doi: 10.1016/S0141-0229(02)00293-4. [DOI] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake P., Wadhwa D. WORLD BANK BLOGS; 2020. 2020 Year in Review: The Impact of COVID-19 in 12 Charts. [Google Scholar]
- Brandelli A., Daroit D.J., Riffel A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010;85:1735–1750. doi: 10.1007/s00253-009-2398-5. [DOI] [PubMed] [Google Scholar]
- Burrell C.J., Howard C.R., Murphy F.A., Burrell C.J., Howard C.R., Murphy F.A. Antiviral chemotherapy, fenner and white’s medical. Virology. 2017:169–183. [Google Scholar]
- Cawley N.X., Huang J., Peng Loh Y., Dhanvantari S. Reference Module in Biomedical Sciences. Elsevier; 2014. Prohormone convertases. [DOI] [Google Scholar]
- Craik C.S., Page M.J., Madison E.L. Proteases as therapeutics. Biochem. J. 2011 doi: 10.1042/BJ20100965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva R.R. Bacterial and fungal proteolytic enzymes: production, catalysis and potential applications. Appl. Biochem. Biotechnol. 2017;183:1–19. doi: 10.1007/s12010-017-2427-2. [DOI] [PubMed] [Google Scholar]
- de Souza P.M., de Assis Bittencourt M.L., Caprara C.C., de Freitas M., de Almeida R.P.C., Silveira D., Fonseca Y.M., Filho E.X.F., Pessoa Junior A., Magalhães P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015;46:337–346. doi: 10.1590/S1517-838246220140359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi F., Marrakchi M., Gargouri M., Saulnier J., Jaffrezic-Renault N., Lagarde F. Comparison of carboxypeptidase y and thermolysin for ochratoxin A electrochemical biosensing. Anal. Methods. 2015 doi: 10.1039/c5ay01905b. [DOI] [Google Scholar]
- El-Fakharany E.M., Haroun B.M., Ng T.B., Redwan E.-R.M. Oyster mushroom laccase inhibits hepatitis C virus entry into peripheral blood cells and hepatoma cells. Protein Pept. Lett. 2010;17:1031–1039. doi: 10.2174/092986610791498948. [DOI] [PubMed] [Google Scholar]
- Enjoji S., Ohama T., Sato K. Regulation of epithelial cell tight junctions by protease-activated receptor 2. J. Vet. Med. Sci. 2014;76:1225–1229. doi: 10.1292/jvms.14-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R.S. Handbook of Proteolytic Enzymes. Elsevier Ltd; 2013. Kexin; pp. 3270–3277. [DOI] [Google Scholar]
- Hauptmann P., Lehle L. Kex1 protease is involved in yeast cell death induced by defective N-glycosylation, acetic acid, and chronological aging. J. Biol. Chem. 2008;283:19151–19163. doi: 10.1074/jbc.M801303200. [DOI] [PubMed] [Google Scholar]
- He X.J., Li X.L., Li Y.Z. Disruption of Cerevisin via Agrobacterium tumefaciens-mediated transformation affects microsclerotia formation and virulence of Verticillium dahliae. Plant Pathol. 2015;64:1157–1167. doi: 10.1111/ppa.12393. [DOI] [Google Scholar]
- Ichishima E. Handbook of Proteolytic Enzymes. Second edition. Elsevier Inc; 2004. Penicillolysin; pp. 784–786. [DOI] [Google Scholar]
- Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016 doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Bond J.S. Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Tejedor D., Clavería-Gimeno R., Velazquez-Campoy A., Abian O., Palomo J.M. 2020. Tyrosinase From Mushroom Agaricus Bisporus as an Inhibitor of the Hepatitis C Virus. bioRxiv 2020.12.23.424187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Kong Y., Yao Q., Zhang B., Leng, Wei F., Bian, Jiao H., Balzarini J., Van Damme E., Bao J.ku. Nebrodeolysin, a novel hemolytic protein from mushroom Pleurotus nebrodensis with apoptosis-inducing and anti-HIV-1 effects. Phytomedicine. 2009;16:198–205. doi: 10.1016/j.phymed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Maeda H., Katase T., Sakai D., Takeuchi M., Kusumoto K.I., Amano H., Ishida H., Abe K., Yamagata Y. A novel non-thermostable deuterolysin from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2016;80:1813–1819. doi: 10.1080/09168451.2016.1166933. [DOI] [PubMed] [Google Scholar]
- Maeda Shingo, Maeda Sadatoshi, Ohno K., Kaji N., Hori M., Fujino Y., Tsujimoto H. Protease-activated receptor-2 induces proinflammatory cytokine and chemokine gene expression in canine keratinocytes. Vet. Immunol. Immunopathol. 2013;153:17–25. doi: 10.1016/j.vetimm.2013.01.018. [DOI] [PubMed] [Google Scholar]
- Magrane M., Consortium U.P. UniProt knowledgebase: a hub of integrated protein data. Database. 2011 doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R.M., Nowak M.A. Coinfection and the evolution of parasite virulence. Proc. R. Soc. B Biol. Sci. 1995;261:209–215. doi: 10.1098/rspb.1995.0138. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen T.L., Holmberg S., Petersen J.G.L. Regulated overproduction and secretion of yeast carboxypeptidase Y. Appl. Microbiol. Biotechnol. 1990 doi: 10.1007/BF00164527. [DOI] [PubMed] [Google Scholar]
- Nisha N.S., Divakaran J. Optimization of alkaline protease production from Bacillus subtilis NS isolated from sea water. Afr. J. Biotechnol. 2014;13:1707–1713. doi: 10.5897/ajb2014.13652. [DOI] [Google Scholar]
- Osmulski P.A., Cropper J., Giletto M., Jones C., Killer C., Jiang S., Tepe J., Chatterjee B., Huang T., Gaczynska M.E. Anticancer applications of allosteric inhibitors of proteasome. J. Clin. Oncol. 2017 doi: 10.1200/jco.2017.35.15_suppl.e23066. [DOI] [Google Scholar]
- Patick A.K., Potts K.E. Protease inhibitors as antiviral agents. Clin. Microbiol. Rev. 1998;11:614–627. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M.N., Beever R.E., Boine B., Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009;10:115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D.J., Choi C. Antiviral bioactive compounds of mushrooms and their antiviral mechanisms: a review. Viruses. 2021;13 doi: 10.3390/v13020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Gat Y., Arya S., Kumar V., Panghal A., Kumar A. A review on microbial alkaline protease: an essential tool for various industrial approaches. Ind. Biotechnol. 2019;15:69–78. doi: 10.1089/ind.2018.0032. [DOI] [Google Scholar]
- Singhal P., Nigam V.K., Vidyarthi A.S. Studies on production, characterization and applications of microbial alkaline proteases. Int. J. Adv. Biotechnol. Res. 2012;3(3):653–669. [Google Scholar]
- Sixt S.U., Dahlmann B. Extracellular, circulating proteasomes and ubiquitin - incidence and relevance. Biochim. Biophys. Acta Mol. basis Dis. 2008 doi: 10.1016/j.bbadis.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Sohar I., Sleat D.E., Lobel P. Handbook of Proteolytic Enzymes. Elsevier Ltd; 2013. Tripeptidyl peptidase I; pp. 3350–3356. [DOI] [Google Scholar]
- Stanke M., Steinkamp R., Waack S., Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004 doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M., Müller R., Gaßen B., Wehrstedt R., Fey P., Karow M.A., Eichinger L., Glöckner G., Noegel A.A. A tripeptidyl peptidase 1 is a binding partner of the Golgi pH regulator (GPHR) in Dictyostelium. Dis. Model. Mech. 2017;10:897–907. doi: 10.1242/dmm.029280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers V. Industrial uses of pepsin [WWW document] Sciencing. 2017 https://sciencing.com/industrial-uses-pepsin-6829146.html (accessed 5.13.21) [Google Scholar]
- Tao C., Chen G., Guo W., Xie M., Ma K., Yan L. A quick guide to diagnosis and treatment of pneumonia with novel coronavirus infections. Her. Med. 2020;39:305–307. [Google Scholar]
- ViralZone . Swiss Inst. Bioinforma; 2020. SARS-Cov-2 genome [WWW Document]https://viralzone.expasy.org/9076 URL. (accessed 5.11.21) [Google Scholar]
- Vorou R.M., Papavassiliou V.G., Tsiodras S. Emerging zoonoses and vector-borne infections affecting humans in Europe. Epidemiol. Infect. 2007;135:1231–1247. doi: 10.1017/S0950268807008527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.X., Ng T.B. Isolation of a novel ubiquitin-like protein from Pleurotus ostreatus mushroom with anti-human immunodeficiency virus, translation-inhibitory, and ribonuclease activities. Biochem. Biophys. Res. Commun. 2000;276:587–593. doi: 10.1006/bbrc.2000.3540. [DOI] [PubMed] [Google Scholar]
- Wang H.X., Ng T.B. Purification of a novel low-molecular-mass laccase with HIV-1 reverse transcriptase inhibitory activity from the mushroom Tricholoma giganteum. Biochem. Biophys. Res. Commun. 2004;315:450–454. doi: 10.1016/j.bbrc.2004.01.064. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang H.X., Ng T.B. A peptide with HIV-1 reverse transcriptase inhibitory activity from the medicinal mushroom Russula paludosa. Peptides. 2007;28:560–565. doi: 10.1016/j.peptides.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material